Genomic Impact of Whaling in North Atlantic Fin Whales
- PMID: 35512360
- PMCID: PMC9113106
- DOI: 10.1093/molbev/msac094
Genomic Impact of Whaling in North Atlantic Fin Whales
Abstract
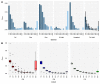
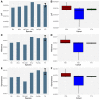
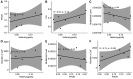
It is generally recognized that large-scale whaling in the 19th and 20th century led to a substantial reduction of the size of many cetacean populations, particularly those of the baleen whales (Mysticeti). The impact of these operations on genomic diversity of one of the most hunted whales, the fin whale (Balaenoptera physalus), has remained largely unaddressed because of the paucity of adequate samples and the limitation of applicable techniques. Here, we have examined the effect of whaling on the North Atlantic fin whale based on genomes of 51 individuals from Icelandic waters, representing three temporally separated intervals, 1989, 2009 and 2018 and provide a reference genome for the species. Demographic models suggest a noticeable drop of the effective population size of the North Atlantic fin whale around a century ago. The present results suggest that the genome-wide heterozygosity is not markedly reduced and has remained comparable with other baleen whale species. Similarly, there are no signs of apparent inbreeding, as measured by the proportion of long runs of homozygosity, or of a distinctively increased mutational load, as measured by the amount of putative deleterious mutations. Compared with other baleen whales, the North Atlantic fin whale appears to be less affected by anthropogenic influences than other whales such as the North Atlantic right whale, consistent with the presence of long runs of homozygosity and higher levels of mutational load in an otherwise more heterozygous genome. Thus, genome-wide assessments of other species and populations are essential for future, more specific, conservation efforts.
Keywords: bottleneck; demography; fin whales; genetic diversity; mutational load; runs of homozygosity; whaling.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Aguilar A, García-Vernet R. 2017. Fin Whale: Balaenoptera physalus. In: Würsig BG, Thewissen JGM, Kovacs KM, editors. Encyclopedia of marine mammals. Amsterdam: Academic Press.
-
- Archer FI, Brownell RL Jr, Hancock-Hanser BL, Morin PA, Robertson KM, Sherman KK, Calambokidis J, Urbán RJ, Rosel PE, Mizroch SA, et al. 2019. Revision of fin whale Balaenoptera physalus (Linnaeus, 1758) subspecies using genetics. Journal of Mammalogy. 100(5):1653–1670.
-
- Árnason Ú. 1981. Fin whales in the NE Atlantic; relationships between abundance and distribution. Ecography. 4(4):245–251.
-
- Bérubé M, Aguilar A, Dendanto D, Larsen F, Di Notarbartolo Sciara G, Sears R, Sigurjónsson J, Urban-R J, Palsbøll PJ. 1998. Population genetic structure of North Atlantic, Mediterranean Sea and Sea of Cortez fin whales, Balaenoptera physalus (Linnaeus 1758). Analysis of mitochondrial and nuclear loci. Mol Ecol. 7(5):585–599. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

