Germline predisposition to pediatric Ewing sarcoma is characterized by inherited pathogenic variants in DNA damage repair genes
- PMID: 35512711
- PMCID: PMC9247831
- DOI: 10.1016/j.ajhg.2022.04.007
Germline predisposition to pediatric Ewing sarcoma is characterized by inherited pathogenic variants in DNA damage repair genes
Abstract
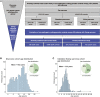
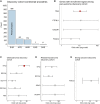
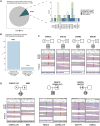
More knowledge is needed regarding germline predisposition to Ewing sarcoma to inform biological investigation and clinical practice. Here, we evaluated the enrichment of pathogenic germline variants in Ewing sarcoma relative to other pediatric sarcoma subtypes, as well as patterns of inheritance of these variants. We carried out European-focused and pan-ancestry case-control analyses to screen for enrichment of pathogenic germline variants in 141 established cancer predisposition genes in 1,147 individuals with pediatric sarcoma diagnoses (226 Ewing sarcoma, 438 osteosarcoma, 180 rhabdomyosarcoma, and 303 other sarcoma) relative to identically processed cancer-free control individuals. Findings in Ewing sarcoma were validated with an additional cohort of 430 individuals, and a subset of 301 Ewing sarcoma parent-proband trios was analyzed for inheritance patterns of identified pathogenic variants. A distinct pattern of pathogenic germline variants was seen in Ewing sarcoma relative to other sarcoma subtypes. FANCC was the only gene with an enrichment signal for heterozygous pathogenic variants in the European Ewing sarcoma discovery cohort (three individuals, OR 12.6, 95% CI 3.0-43.2, p = 0.003, FDR = 0.40). This enrichment in FANCC heterozygous pathogenic variants was again observed in the European Ewing sarcoma validation cohort (three individuals, OR 7.0, 95% CI 1.7-23.6, p = 0.014), representing a broader importance of genes involved in DNA damage repair, which were also nominally enriched in individuals with Ewing sarcoma. Pathogenic variants in DNA damage repair genes were acquired through autosomal inheritance. Our study provides new insight into germline risk factors contributing to Ewing sarcoma pathogenesis.
Keywords: Ewing sarcoma, cancer predisposition, pediatric oncology, genetic risk.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.G. has equity in Google, Microsoft, Amazon, Apple, Moderna, Pfizer, and Vertex Pharmaceuticals. J.K.'s spouse has received consulting fees from ROME Therapeutics, Foundation Medicine, Inc., NanoString Technologies, EMD Millipore Sigma, Pfizer, and Tekla Capital; is a founder and has equity in ROME Therapeutics, PanTher Therapeutics, and TellBio, Inc.; and receives research support from ACD-Biotechne, PureTech Health LLC, and Ribon Therapeutics. J.D.S. is co-founder and shareholder in ItRunsInMyFamily.com and is co-founder, shareholder, and employed by Peel Therapeutics, Inc. E.M.V.A. holds consulting roles with Tango Therapeutics, Genome Medical, Genomic Life, Enara Bio, Janssen, and Manifold Bio; he receives research support from Bristol-Myers Squibb and Novartis; he has equity in Tango Therapeutics, Genome Medical, Genomic Life, Syapse, Enara Bio, Manifold Bio, and Microsoft; he has received travel reimbursement from Roche and Genentech; and he has filed institutional patents on chromatin mutations, immunotherapy response, and methods for clinical interpretation. The other authors declare no competing interests.
Figures

Similar articles
-
Frequent inactivating germline mutations in DNA repair genes in patients with Ewing sarcoma.Genet Med. 2017 Aug;19(8):955-958. doi: 10.1038/gim.2016.206. Epub 2017 Jan 26. Genet Med. 2017. PMID: 28125078 Free PMC article.
-
Germline variants in patients diagnosed with pediatric soft tissue sarcoma.Acta Oncol. 2024 Jul 22;63:586-591. doi: 10.2340/1651-226X.2024.40730. Acta Oncol. 2024. PMID: 39037077 Free PMC article.
-
Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients With Osteosarcoma.JAMA Oncol. 2020 May 1;6(5):724-734. doi: 10.1001/jamaoncol.2020.0197. JAMA Oncol. 2020. PMID: 32191290 Free PMC article.
-
Germline Variation and Somatic Alterations in Ewing Sarcoma.Methods Mol Biol. 2021;2226:3-14. doi: 10.1007/978-1-0716-1020-6_1. Methods Mol Biol. 2021. PMID: 33326089 Review.
-
Investigation of clinically relevant germline variants detected by next-generation sequencing in patients with childhood cancer: a review of the literature.J Med Genet. 2018 Dec;55(12):785-793. doi: 10.1136/jmedgenet-2018-105488. Epub 2018 Oct 4. J Med Genet. 2018. PMID: 30287599 Review.
Cited by
-
Small round cell sarcomas.Nat Rev Dis Primers. 2022 Oct 6;8(1):66. doi: 10.1038/s41572-022-00393-3. Nat Rev Dis Primers. 2022. PMID: 36202860 Review.
-
BARD1 germline variants induce haploinsufficiency and DNA repair defects in neuroblastoma.bioRxiv [Preprint]. 2023 Feb 3:2023.01.31.525066. doi: 10.1101/2023.01.31.525066. bioRxiv. 2023. Update in: J Natl Cancer Inst. 2024 Jan 10;116(1):138-148. doi: 10.1093/jnci/djad182. PMID: 36778420 Free PMC article. Updated. Preprint.
-
Genetic Predisposition to Sarcoma: What Should Clinicians Know?Curr Treat Options Oncol. 2024 Jun;25(6):769-783. doi: 10.1007/s11864-024-01192-6. Epub 2024 May 7. Curr Treat Options Oncol. 2024. PMID: 38713268 Review.
-
Most Fanconi anemia heterozygotes are not at increased cancer risk: A genome-first DiscovEHR cohort population study.Genet Med. 2024 Mar;26(3):101042. doi: 10.1016/j.gim.2023.101042. Epub 2023 Dec 5. Genet Med. 2024. PMID: 38063144 Free PMC article.
-
NCI Cancer Research Data Commons: Cloud-Based Analytic Resources.Cancer Res. 2024 May 2;84(9):1396-1403. doi: 10.1158/0008-5472.CAN-23-2657. Cancer Res. 2024. PMID: 38488504 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

