Epigenetic changes induced by in utero dietary challenge result in phenotypic variability in successive generations of mice
- PMID: 35513363
- PMCID: PMC9072353
- DOI: 10.1038/s41467-022-30022-2
Epigenetic changes induced by in utero dietary challenge result in phenotypic variability in successive generations of mice
Abstract
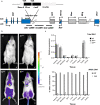
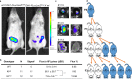
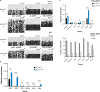
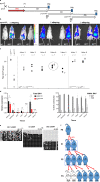
Transmission of epigenetic information between generations occurs in nematodes, flies and plants, mediated by specialised small RNA pathways, modified histones and DNA methylation. Similar processes in mammals can also affect phenotype through intergenerational or trans-generational mechanisms. Here we generate a luciferase knock-in reporter mouse for the imprinted Dlk1 locus to visualise and track epigenetic fidelity across generations. Exposure to high-fat diet in pregnancy provokes sustained re-expression of the normally silent maternal Dlk1 in offspring (loss of imprinting) and increased DNA methylation at the somatic differentially methylated region (sDMR). In the next generation heterogeneous Dlk1 mis-expression is seen exclusively among animals born to F1-exposed females. Oocytes from these females show altered gene and microRNA expression without changes in DNA methylation, and correct imprinting is restored in subsequent generations. Our results illustrate how diet impacts the foetal epigenome, disturbing canonical and non-canonical imprinting mechanisms to modulate the properties of successive generations of offspring.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Samata, M., et al. Intergenerationally Maintained Histone H4 Lysine 16 Acetylation Is Instructive for Future Gene Activation. Cell. 182, 127–144.e23 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
- PS3125_WCMA/WT_/Wellcome Trust/United Kingdom
- MC_UP_1605/11/MRC_/Medical Research Council/United Kingdom
- BB/P008623/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/S000437/1/MRC_/Medical Research Council/United Kingdom
- MC_U120027516/MRC_/Medical Research Council/United Kingdom
- 099276/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- BB/P002307/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UP_1605/12/MRC_/Medical Research Council/United Kingdom
- MC_UP_A652_1001/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/000C0423/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

