Small extracellular vesicle-mediated miR-320e transmission promotes osteogenesis in OPLL by targeting TAK1
- PMID: 35513391
- PMCID: PMC9072352
- DOI: 10.1038/s41467-022-29029-6
Small extracellular vesicle-mediated miR-320e transmission promotes osteogenesis in OPLL by targeting TAK1
Abstract
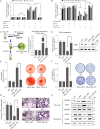
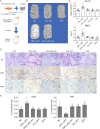
Ossification of the posterior longitudinal ligament (OPLL) is an emerging spinal disease caused by heterotopic ossification of the posterior longitudinal ligament. The pathological mechanism is poorly understood, which hinders the development of nonsurgical treatments. Here, we set out to explore the function and mechanism of small extracellular vesicles (sEVs) in OPLL. Global miRNA sequencings are performed on sEVs derived from ligament cells of normal and OPLL patients, and we have showed that miR-320e is abundantly expressed in OPLL-derived sEVs compare to other sEVs. Treatment with either sEVs or miR-320e significantly promote the osteoblastic differentiation of normal longitudinal ligament cells and mesenchymal stem cells and inhibit the osteoclastic differentiation of monocytes. Through a mechanistic study, we find that TAK1 is a downstream target of miR-320e, and we further validate these findings in vivo using OPLL model mice. Together, our data demonstrate that OPLL ligament cells secrete ossification-promoting sEVs that contribute to the development of ossification through the miR-320e/TAK1 axis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
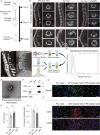
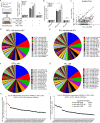
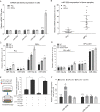

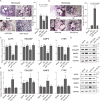



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

