Combinatorial GxGxE CRISPR screen identifies SLC25A39 in mitochondrial glutathione transport linking iron homeostasis to OXPHOS
- PMID: 35513392
- PMCID: PMC9072411
- DOI: 10.1038/s41467-022-30126-9
Combinatorial GxGxE CRISPR screen identifies SLC25A39 in mitochondrial glutathione transport linking iron homeostasis to OXPHOS
Abstract
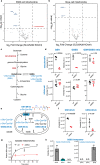
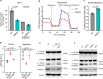
The SLC25 carrier family consists of 53 transporters that shuttle nutrients and co-factors across mitochondrial membranes. The family is highly redundant and their transport activities coupled to metabolic state. Here, we use a pooled, dual CRISPR screening strategy that knocks out pairs of transporters in four metabolic states - glucose, galactose, OXPHOS inhibition, and absence of pyruvate - designed to unmask the inter-dependence of these genes. In total, we screen 63 genes in four metabolic states, corresponding to 2016 single and pair-wise genetic perturbations. We recover 19 gene-by-environment (GxE) interactions and 9 gene-by-gene (GxG) interactions. One GxE interaction hit illustrates that the fitness defect in the mitochondrial folate carrier (SLC25A32) KO cells is genetically buffered in galactose due to a lack of substrate in de novo purine biosynthesis. GxG analysis highlights a buffering interaction between the iron transporter SLC25A37 (A37) and the poorly characterized SLC25A39 (A39). Mitochondrial metabolite profiling, organelle transport assays, and structure-guided mutagenesis identify A39 as critical for mitochondrial glutathione (GSH) import. Functional studies reveal that A39-mediated glutathione homeostasis and A37-mediated mitochondrial iron uptake operate jointly to support mitochondrial OXPHOS. Our work underscores the value of studying family-wide genetic interactions across different metabolic environments.
© 2022. The Author(s).
Conflict of interest statement
V.K.M. serves on the SAB and receives equity from Janssen Pharmaceutics and 5AM Ventures. J.G.D. consults for Foghorn Therapeutics, Maze Therapeutics, Merck, Agios, and Pfizer; J.G.D. consults for and has equity in Tango Therapeutics. The remaining authors declare no competing interests.
Figures




Similar articles
-
Dual regulation of SLC25A39 by AFG3L2 and iron controls mitochondrial glutathione homeostasis.Mol Cell. 2024 Feb 15;84(4):802-810.e6. doi: 10.1016/j.molcel.2023.12.008. Epub 2023 Dec 28. Mol Cell. 2024. PMID: 38157846 Free PMC article.
-
SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells.Nature. 2021 Nov;599(7883):136-140. doi: 10.1038/s41586-021-04025-w. Epub 2021 Oct 27. Nature. 2021. PMID: 34707288 Free PMC article.
-
Autoregulatory control of mitochondrial glutathione homeostasis.Science. 2023 Nov 17;382(6672):820-828. doi: 10.1126/science.adf4154. Epub 2023 Nov 2. Science. 2023. PMID: 37917749 Free PMC article.
-
SLC25A39 links mitochondrial GSH sensing with iron metabolism.Mol Cell. 2024 Feb 15;84(4):616-618. doi: 10.1016/j.molcel.2023.12.037. Mol Cell. 2024. PMID: 38364779 Review.
-
Techniques for investigating mitochondrial gene expression.BMB Rep. 2020 Jan;53(1):3-9. doi: 10.5483/BMBRep.2020.53.1.272. BMB Rep. 2020. PMID: 31818361 Free PMC article. Review.
Cited by
-
Metabolic determinants of tumour initiation.Nat Rev Endocrinol. 2023 Mar;19(3):134-150. doi: 10.1038/s41574-022-00773-5. Epub 2022 Nov 29. Nat Rev Endocrinol. 2023. PMID: 36446897 Free PMC article. Review.
-
Iron and Cancer.Adv Exp Med Biol. 2025;1480:271-289. doi: 10.1007/978-3-031-92033-2_18. Adv Exp Med Biol. 2025. PMID: 40603797 Review.
-
JIP4 deficiency causes a lysosomal storage disease arising from impaired cystine efflux.bioRxiv [Preprint]. 2025 Jun 8:2025.06.06.657909. doi: 10.1101/2025.06.06.657909. bioRxiv. 2025. PMID: 40661466 Free PMC article. Preprint.
-
Nutrient transporters: connecting cancer metabolism to therapeutic opportunities.Oncogene. 2023 Mar;42(10):711-724. doi: 10.1038/s41388-023-02593-x. Epub 2023 Feb 4. Oncogene. 2023. PMID: 36739364 Free PMC article. Review.
-
Mitoferrin2 is a synthetic lethal target for chromosome 8p deleted cancers.Genome Med. 2024 Jun 17;16(1):83. doi: 10.1186/s13073-024-01357-w. Genome Med. 2024. PMID: 38886830 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

