PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements
- PMID: 35513404
- PMCID: PMC9072354
- DOI: 10.1038/s41467-022-30168-z
PICASSO allows ultra-multiplexed fluorescence imaging of spatially overlapping proteins without reference spectra measurements
Abstract
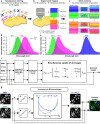
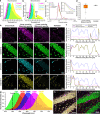
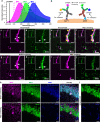
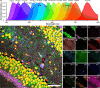
Ultra-multiplexed fluorescence imaging requires the use of spectrally overlapping fluorophores to label proteins and then to unmix the images of the fluorophores. However, doing this remains a challenge, especially in highly heterogeneous specimens, such as the brain, owing to the high degree of variation in the emission spectra of fluorophores in such specimens. Here, we propose PICASSO, which enables more than 15-color imaging of spatially overlapping proteins in a single imaging round without using any reference emission spectra. PICASSO requires an equal number of images and fluorophores, which enables such advanced multiplexed imaging, even with bandpass filter-based microscopy. We show that PICASSO can be used to achieve strong multiplexing capability in diverse applications. By combining PICASSO with cyclic immunofluorescence staining, we achieve 45-color imaging of the mouse brain in three cycles. PICASSO provides a tool for multiplexed imaging with high accessibility and accuracy for a broad range of researchers.
© 2022. The Author(s).
Conflict of interest statement
I.C. and H.N. declare no competing interests. J.-B.C., Y.-G.Y., J.S., Y.S., H.K., and J.K. declare the following competing interests. J.-B.C., Y.-G.Y., J.S., Y.S., H.K., and J.K. are co-inventors on patent applications owned by KAIST covering PICASSO (KR10-2020-0088091, KR10-2021-0089419, KR10-2021-0090057, KR10-2021-0171431, US 17/132628, US17/445629, US17/567300, PCT/KR2021/011097, PCT/KR2021/018833, EP22150031.7, EP21192162.2).
Figures


Comment in
-
Unmixing for ultra-high-plex fluorescence imaging.Nat Commun. 2022 Jun 16;13(1):3473. doi: 10.1038/s41467-022-31110-z. Nat Commun. 2022. PMID: 35710800 Free PMC article.
References
-
- Zimmermann, T., Marrison, J., Hogg, K. & O’Toole, P. In Confocal Microscopy: Methods and Protocols (ed. Paddock, S. W.) 129–148 (Springer New York, 2014).
-
- Zimmermann, T., Rietdorf, J. & Pepperkok, R. Spectral imaging and its applications in live cell microscopy. FEBS Lett. 10.1016/S0014-5793(03)00521-0 (2003). - PubMed
-
- Gorris, M. A. J. et al. Eight-color multiplex immunohistochemistry for simultaneous detection of multiple immune checkpoint molecules within the tumor microenvironment. J. Immunol. 10.4049/jimmunol.1701262 (2018). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

