Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease
- PMID: 35513515
- PMCID: PMC9076534
- DOI: 10.1038/s41593-022-01061-1
Single-cell genomic profiling of human dopamine neurons identifies a population that selectively degenerates in Parkinson's disease
Abstract
The loss of dopamine (DA) neurons within the substantia nigra pars compacta (SNpc) is a defining pathological hallmark of Parkinson's disease (PD). Nevertheless, the molecular features associated with DA neuron vulnerability have not yet been fully identified. Here, we developed a protocol to enrich and transcriptionally profile DA neurons from patients with PD and matched controls, sampling a total of 387,483 nuclei, including 22,048 DA neuron profiles. We identified ten populations and spatially localized each within the SNpc using Slide-seq. A single subtype, marked by the expression of the gene AGTR1 and spatially confined to the ventral tier of SNpc, was highly susceptible to loss in PD and showed the strongest upregulation of targets of TP53 and NR2F2, nominating molecular processes associated with degeneration. This same vulnerable population was specifically enriched for the heritable risk associated with PD, highlighting the importance of cell-intrinsic processes in determining the differential vulnerability of DA neurons to PD-associated degeneration.
© 2022. The Author(s).
Conflict of interest statement
E.Z.M. is a consultant for Atlas Bio, Inc. The remaining authors declare no competing interests.
Figures









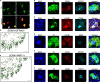


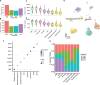

Comment in
-
Parkinson's disease in the single-cell era.Nat Neurosci. 2022 May;25(5):536-538. doi: 10.1038/s41593-022-01069-7. Nat Neurosci. 2022. PMID: 35513514 No abstract available.
-
Vulnerable dopamine neurons identified.Nat Rev Neurol. 2022 Jul;18(7):381. doi: 10.1038/s41582-022-00682-1. Nat Rev Neurol. 2022. PMID: 35676370 No abstract available.
-
Nigral Neurons Degenerating in Parkinson's Disease Express the Angiotensin Receptor Type 1 Gene.Mov Disord. 2022 Aug;37(8):1610-1611. doi: 10.1002/mds.29137. Epub 2022 Jun 29. Mov Disord. 2022. PMID: 35766438 Free PMC article.
References
-
- Bloem, B. R., Okun, M. S. & Klein, C. Parkinson’s disease. Lancet397, P2284–P2303 (2021). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

