Ambient mass spectrometry for rapid authentication of milk from Alpine or lowland forage
- PMID: 35513691
- PMCID: PMC9072378
- DOI: 10.1038/s41598-022-11178-9
Ambient mass spectrometry for rapid authentication of milk from Alpine or lowland forage
Abstract
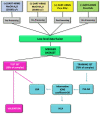
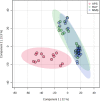
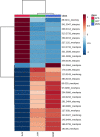
Metabolomics approaches, such as direct analysis in real time-high resolution mass spectrometry (DART-HRMS), allow characterising many polar and non-polar compounds useful as authentication biomarkers of dairy chains. By using both a partial least squares discriminant analysis (PLS-DA) and a linear discriminant analysis (LDA), this study aimed to assess the capability of DART-HRMS, coupled with a low-level data fusion, discriminate among milk samples from lowland (silages vs. hay) and Alpine (grazing; APS) systems and identify the most informative biomarkers associated with the main dietary forage. As confirmed also by the LDA performed against the test set, DART-HRMS analysis provided an accurate discrimination of Alpine samples; meanwhile, there was a limited capacity to correctly recognise silage- vs. hay-milks. Supervised multivariate statistics followed by metabolomics hierarchical cluster analysis allowed extrapolating the most significant metabolites. Lowland milk was characterised by a pool of energetic compounds, ketoacid derivates, amines and organic acids. Seven informative DART-HRMS molecular features, mainly monoacylglycerols, could strongly explain the metabolomic variation of Alpine grazing milk and contributed to its classification. The misclassification between the two lowland groups confirmed that the intensive dairy systems would be characterised by a small variation in milk composition.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Prediction of indicators of cow diet composition and authentication of feeding specifications of Protected Designation of Origin cheese using mid-infrared spectroscopy on milk.J Dairy Sci. 2021 Jan;104(1):112-125. doi: 10.3168/jds.2020-18468. Epub 2020 Nov 6. J Dairy Sci. 2021. PMID: 33162089
-
Changes of Milk Metabolomic Profiles Resulting from a Mycotoxins-Contaminated Corn Silage Intake by Dairy Cows.Metabolites. 2021 Jul 23;11(8):475. doi: 10.3390/metabo11080475. Metabolites. 2021. PMID: 34436416 Free PMC article.
-
Evaluation of direct analysis in real time ionization-mass spectrometry (DART-MS) in fish metabolomics aimed to assess the response to dietary supplementation.Talanta. 2013 Oct 15;115:263-70. doi: 10.1016/j.talanta.2013.04.025. Epub 2013 Apr 15. Talanta. 2013. PMID: 24054590
-
Milk metabolomics based on ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry to discriminate different cows feeding regimens.Food Res Int. 2020 Aug;134:109279. doi: 10.1016/j.foodres.2020.109279. Epub 2020 Apr 28. Food Res Int. 2020. PMID: 32517942
-
Nutritive value of maize silage in relation to dairy cow performance and milk quality.J Sci Food Agric. 2015 Jan;95(2):238-52. doi: 10.1002/jsfa.6703. Epub 2014 Jun 2. J Sci Food Agric. 2015. PMID: 24752455 Review.
Cited by
-
A Comprehensive Review of Milk Components: Recent Developments on Extraction and Analysis Methods.Molecules. 2025 Apr 30;30(9):1994. doi: 10.3390/molecules30091994. Molecules. 2025. PMID: 40363801 Free PMC article. Review.
-
A Snapshot, Using a Multi-Omic Approach, of the Metabolic Cross-Talk and the Dynamics of the Resident Microbiota in Ripening Cheese Inoculated with Listeria innocua.Foods. 2024 Jun 18;13(12):1912. doi: 10.3390/foods13121912. Foods. 2024. PMID: 38928853 Free PMC article.
-
Metabolic signature of Mycobacterium avium subsp. paratuberculosis infected and infectious dairy cattle by integrating nuclear magnetic resonance analysis and blood indices.Front Vet Sci. 2023 Apr 17;10:1146626. doi: 10.3389/fvets.2023.1146626. eCollection 2023. Front Vet Sci. 2023. PMID: 37138915 Free PMC article.
-
Effect of Diet on CPFAs Used as Markers in Milk for the Detection of Silage in the Ration of Dairy Cows.Foods. 2025 Feb 2;14(3):476. doi: 10.3390/foods14030476. Foods. 2025. PMID: 39942069 Free PMC article.
-
Rapid, novel screening of toxicants in poison baits, and autopsy specimens by ambient mass spectrometry.Front Chem. 2022 Aug 25;10:982377. doi: 10.3389/fchem.2022.982377. eCollection 2022. Front Chem. 2022. PMID: 36092679 Free PMC article.
References
-
- Lanza I, et al. Use of GC-MS and 1H NMR low-level data fusion as an advanced and comprehensive metabolomic approach to discriminate milk from dairy chains based on different types of forage. Int. Dairy J. 2021;123:105174. doi: 10.1016/j.idairyj.2021.105174. - DOI
-
- Bär C, et al. Impact of herbage proportion, animal breed, lactation stage and season on the fatty acid and protein composition of milk. Int. Dairy J. 2020;109:104785. doi: 10.1016/j.idairyj.2020.104785. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

