Gene silencing, knockout and over-expression of a transcription factor ABORTED MICROSPORES (SlAMS) strongly affects pollen viability in tomato (Solanum lycopersicum)
- PMID: 35513810
- PMCID: PMC9069838
- DOI: 10.1186/s12864-022-08549-x
Gene silencing, knockout and over-expression of a transcription factor ABORTED MICROSPORES (SlAMS) strongly affects pollen viability in tomato (Solanum lycopersicum)
Abstract
Background: The tomato (Solanum lycopersicum L.) is an economically valuable crop grown worldwide. Because the use of sterile males reduces the cost of F1 seed production, the innovation of male sterility is of great significance for tomato breeding. The ABORTED MICROSPORES gene (AMS), which encodes for a basic helix-loop-helix (bHLH) transcription factor, has been previously indicated as an essential gene for tapetum development in Arabidopsis and rice. To determine the function of the SlAMS gene (AMS gene from S. lycopersicum) and verify whether it is a potential candidate gene for generating the male sterility in tomato, we used virus-induced gene silencing (VIGS), CRISPR/Cas9-mediated genome editing and over-expression technology to transform tomato via Agrobacterium infection.
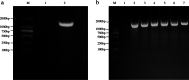
Results: Here, the full-length SlAMS gene with 1806 bp from S. lycopersicum (Accession No. MK591950.1) was cloned from pollen cDNA. The results of pollen grains staining showed that, the non-viable pollen proportions of SlAMS-silenced (75%), -knockouted (89%) and -overexpressed plants (60%) were significantly higher than the wild type plants (less than 10%; P < 0.01). In three cases, the morphology of non-viable pollen grains appeared tetragonal, circular, atrophic, shriveled, or otherwise abnormally shaped, while those of wild type appeared oval and plump. Furthermore, the qRT-PCR analysis indicated that SlAMS in anthers of SlAMS-silenced and -knockouted plants had remarkably lower expression than in that of wild type (P < 0.01), and yet it had higher expression in SlAMS-overexpressed plants (P < 0.01).
Conclusion: In this paper, Our research suggested alternative approaches to generating male sterility in tomato, among which CRISPR/Cas9-mediated editing of SlAMS implied the best performance. We also demonstrated that the downregulation and upregulation of SlAMS both affected the pollen formation and notably led to reduction of pollen viability, suggesting SlAMS might be essential for regulating pollen development in tomato. These findings may facilitate studies on clarifying the SlAMS-associated molecular regulatory mechanism of pollen development in tomato.
Keywords: Gene knockout; Over-expression; SlAMS; Tomato; VIGS.
© 2022. The Author(s).
Conflict of interest statement
The authors declared that they have no conflict of interests.
Figures



References
-
- Radkova M, Balacheva E, Atanassova B, Iantcheva A, Atanassov A. Study on the Potential of Genic Male Sterility in Tomato as a Tool for Pollen Flow Restriction. Biotechnol Biotechnol Equip. 2014;23(3):1303–1308. doi: 10.1080/13102818.2009.10817658. - DOI
-
- Bhatia P, Ashwath N, Senaratna T, Midmore D. Tissue Culture Studies of Tomato (Lycopersicon esculentum) PCTOC. 2004;78(1):1–21. doi: 10.1023/B:TICU.0000020430.08558.6e. - DOI
-
- Atanassova B. Functional male sterility in tomato (Lycopersicon esculentum Mill.) and its application in hybrid seed production. Acta Physiologiae Plantarum. 2000;22(3):221–225. doi: 10.1007/s11738-000-0015-4. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

