In silico and in vitro metabolism of ribociclib: a mass spectrometric approach to bioactivation pathway elucidation and metabolite profiling
- PMID: 35514564
- PMCID: PMC9054585
- DOI: 10.1039/d0ra01624a
In silico and in vitro metabolism of ribociclib: a mass spectrometric approach to bioactivation pathway elucidation and metabolite profiling
Erratum in
-
Correction: In silico and in vitro metabolism of ribociclib: a mass spectrometric approach to bioactivation pathway elucidation and metabolite profiling.RSC Adv. 2020 Jun 23;10(40):23930. doi: 10.1039/d0ra90070b. eCollection 2020 Jun 19. RSC Adv. 2020. PMID: 35532509 Free PMC article.
Abstract
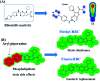
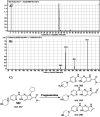
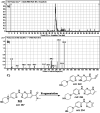
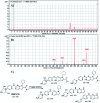
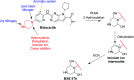
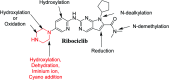
Ribociclib (RBC, Kisqali®) is a highly selective CDK4/6 inhibitor that has been approved for breast cancer therapy. Initially, prediction of susceptible sites of metabolism and reactivity pathways were performed by the StarDrop WhichP450™ module and the Xenosite web predictor tool, respectively. Later, in vitro metabolites and adducts of RBC were characterized from rat liver microsomes using LC-MS/MS. Subsequently, in silico data was used as a guide for the in vitro work. Finally, in silico toxicity assessment of RBC metabolites was carried out using DEREK software and structural modification was proposed to reduce their side effects and to validate the bioactivation pathway theory using the StarDrop DEREK module. In vitro phase I metabolic profiling of RBC was performed utilizing rat liver microsomes (RLMs). Generation of reactive metabolites was investigated using potassium cyanide (KCN) as a trapping nucleophile for the transient and reactive iminium intermediates to form a stable cyano adduct that can be identified and characterized using mass spectrometry. Nine phase I metabolites and one cyano adduct of RBC were characterized. The proposed metabolic pathways involved in generation of these metabolites are hydroxylation, oxidation and reduction. The reactive intermediate generation mechanism of RBC may provide an explanation of its adverse reactions. Aryl piperazine is considered a structural alert for toxicity as proposed by the DEREK report. We propose that the generation of only one reactive metabolite of RBC in a very small concentration is due to the decreased reactivity of the piperazine ring compared to previous reports of similar drugs. Docking analysis was performed for RBC and its proposed derivatives at the active site of the human CDK6 enzyme. Methyl-RBC exhibited the best ADMET and docking analysis and fewer side effects compared to RBC and fluoro-RBC. Further drug discovery studies can be conducted taking into account this concept allowing the development of new drugs with enhanced safety profiles that were confirmed by using StarDrop software. To the best of our knowledge, this is the first literature report of RBCin vitro metabolic profiling and structural characterization and toxicological properties of the generated metabolites.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- American cancer society,http://www.cancer.org/acs/groups/cid/documents/webcontent/003090-pdf.pdf
-
- National Campaign for Breast Cancer Awareness, http://www.moh.gov.sa/en/HealthAwareness/Campaigns/Breastcancer/Pages/st...
LinkOut - more resources
Full Text Sources

