E Protein Transcription Factors as Suppressors of T Lymphocyte Acute Lymphoblastic Leukemia
- PMID: 35514954
- PMCID: PMC9065262
- DOI: 10.3389/fimmu.2022.885144
E Protein Transcription Factors as Suppressors of T Lymphocyte Acute Lymphoblastic Leukemia
Abstract
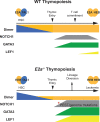
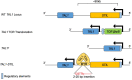
T Lymphocyte Acute Lymphoblastic Leukemia (ALL) is an aggressive disease arising from transformation of T lymphocytes during their development. The mutation spectrum of T-ALL has revealed critical regulators of the growth and differentiation of normal and leukemic T lymphocytes. Approximately, 60% of T-ALLs show aberrant expression of the hematopoietic stem cell-associated helix-loop-helix transcription factors TAL1 and LYL1. TAL1 and LYL1 function in multiprotein complexes that regulate gene expression in T-ALL but they also antagonize the function of the E protein homodimers that are critical regulators of T cell development. Mice lacking E2A, or ectopically expressing TAL1, LYL1, or other inhibitors of E protein function in T cell progenitors, also succumb to an aggressive T-ALL-like disease highlighting that E proteins promote T cell development and suppress leukemogenesis. In this review, we discuss the role of E2A in T cell development and how alterations in E protein function underlie leukemogenesis. We focus on the role of TAL1 and LYL1 and the genes that are dysregulated in E2a-/- T cell progenitors that contribute to human T-ALL. These studies reveal novel mechanisms of transformation and provide insights into potential therapeutic targets for intervention in this disease.
Keywords: E protein; LYL1; Leukemia; T lymphocyte; TAL1; murine.
Copyright © 2022 Parriott and Kee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Loss of thymocyte competition underlies the tumor suppressive functions of the E2a transcription factor in T-ALL.Leukemia. 2024 Mar;38(3):491-501. doi: 10.1038/s41375-023-02123-4. Epub 2023 Dec 28. Leukemia. 2024. PMID: 38155245
-
Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL.Blood. 2013 Sep 19;122(12):2093-103. doi: 10.1182/blood-2012-09-458570. Epub 2013 Aug 7. Blood. 2013. PMID: 23926305
-
Multiple mechanisms induce ectopic expression of LYL1 in subsets of T-ALL cell lines.Leuk Res. 2010 Apr;34(4):521-8. doi: 10.1016/j.leukres.2009.06.020. Epub 2009 Jul 15. Leuk Res. 2010. PMID: 19608273
-
TAL1, TAL2 and LYL1: a family of basic helix-loop-helix proteins implicated in T cell acute leukaemia.Semin Cancer Biol. 1993 Dec;4(6):341-7. Semin Cancer Biol. 1993. PMID: 8142619 Review.
-
Concise review: Blood relatives: formation and regulation of hematopoietic stem cells by the basic helix-loop-helix transcription factors stem cell leukemia and lymphoblastic leukemia-derived sequence 1.Stem Cells. 2012 Jun;30(6):1053-8. doi: 10.1002/stem.1093. Stem Cells. 2012. PMID: 22593015 Review.
Cited by
-
Ubiquitin-specific proteases (USPs) in leukemia: a systematic review.BMC Cancer. 2024 Jul 25;24(1):894. doi: 10.1186/s12885-024-12614-x. BMC Cancer. 2024. PMID: 39048945 Free PMC article.
-
The emerging applications and advancements of Raman spectroscopy in pediatric cancers.Front Oncol. 2023 Feb 6;13:1044177. doi: 10.3389/fonc.2023.1044177. eCollection 2023. Front Oncol. 2023. PMID: 36814817 Free PMC article. Review.
-
Loss of thymocyte competition underlies the tumor suppressive functions of the E2a transcription factor in T-ALL.Leukemia. 2024 Mar;38(3):491-501. doi: 10.1038/s41375-023-02123-4. Epub 2023 Dec 28. Leukemia. 2024. PMID: 38155245
-
Transcription Factor TAL1 in Erythropoiesis.Adv Exp Med Biol. 2024;1459:243-258. doi: 10.1007/978-3-031-62731-6_11. Adv Exp Med Biol. 2024. PMID: 39017847 Review.
-
Multi-modular structure of the gene regulatory network for specification and commitment of murine T cells.Front Immunol. 2023 Jan 31;14:1108368. doi: 10.3389/fimmu.2023.1108368. eCollection 2023. Front Immunol. 2023. PMID: 36817475 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

