Inflammatory Immune-Associated eRNA: Mechanisms, Functions and Therapeutic Prospects
- PMID: 35514959
- PMCID: PMC9063412
- DOI: 10.3389/fimmu.2022.849451
Inflammatory Immune-Associated eRNA: Mechanisms, Functions and Therapeutic Prospects
Abstract
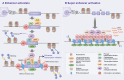
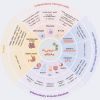
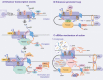
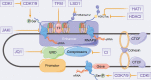
The rapid development of multiple high-throughput sequencing technologies has made it possible to explore the critical roles and mechanisms of functional enhancers and enhancer RNAs (eRNAs). The inflammatory immune response, as a fundamental pathological process in infectious diseases, cancers and immune disorders, coordinates the balance between the internal and external environment of the organism. It has been shown that both active enhancers and intranuclear eRNAs are preferentially expressed over inflammation-related genes in response to inflammatory stimuli, suggesting that enhancer transcription events and their products influence the expression and function of inflammatory genes. Therefore, in this review, we summarize and discuss the relevant inflammatory roles and regulatory mechanisms of eRNAs in inflammatory immune cells, non-inflammatory immune cells, inflammatory immune diseases and tumors, and explore the potential therapeutic effects of enhancer inhibitors affecting eRNA production for diseases with inflammatory immune responses.
Keywords: cancers; eRNA; enhancer transcription events; immune inflammatory; therapeutic prospects.
Copyright © 2022 Wan, Li, Meng, Hou, Chen and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Enhancer RNAs step forward: new insights into enhancer function.Development. 2022 Aug 15;149(16):dev200398. doi: 10.1242/dev.200398. Epub 2022 Aug 30. Development. 2022. PMID: 36039999 Free PMC article.
-
Enhancer RNA (eRNA) in Human Diseases.Int J Mol Sci. 2022 Sep 30;23(19):11582. doi: 10.3390/ijms231911582. Int J Mol Sci. 2022. PMID: 36232885 Free PMC article. Review.
-
Genetic, Pharmacogenomic, and Immune Landscapes of Enhancer RNAs Across Human Cancers.Cancer Res. 2022 Mar 1;82(5):785-790. doi: 10.1158/0008-5472.CAN-21-2058. Cancer Res. 2022. PMID: 35022213
-
Enhancer RNA: What we know and what we can achieve.Cell Prolif. 2022 Apr;55(4):e13202. doi: 10.1111/cpr.13202. Epub 2022 Feb 16. Cell Prolif. 2022. PMID: 35170113 Free PMC article. Review.
-
Pig-eRNAdb: a comprehensive enhancer and eRNA dataset of pigs.Sci Data. 2024 Feb 1;11(1):157. doi: 10.1038/s41597-024-02960-7. Sci Data. 2024. PMID: 38302497 Free PMC article.
Cited by
-
LncRNAs, nuclear architecture and the immune response.Nucleus. 2024 Dec;15(1):2350182. doi: 10.1080/19491034.2024.2350182. Epub 2024 May 13. Nucleus. 2024. PMID: 38738760 Free PMC article. Review.
-
Enhancer RNA in cancer: identification, expression, resources, relationship with immunity, drugs, and prognosis.Brief Funct Genomics. 2025 Jan 15;24:elaf007. doi: 10.1093/bfgp/elaf007. Brief Funct Genomics. 2025. PMID: 40285345 Free PMC article. Review.
-
Amplifying gene expression with RNA-targeted therapeutics.Nat Rev Drug Discov. 2023 Jul;22(7):539-561. doi: 10.1038/s41573-023-00704-7. Epub 2023 May 30. Nat Rev Drug Discov. 2023. PMID: 37253858 Free PMC article. Review.
-
Accelerating CAR-T Cell Therapies with Small-Molecule Inhibitors.BioDrugs. 2025 Jan;39(1):33-51. doi: 10.1007/s40259-024-00688-9. Epub 2024 Nov 26. BioDrugs. 2025. PMID: 39589646 Free PMC article. Review.
-
Non-Coding RNAs and Innate Immune Responses in Cancer.Biomedicines. 2024 Sep 11;12(9):2072. doi: 10.3390/biomedicines12092072. Biomedicines. 2024. PMID: 39335585 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

