Serum Antigenome Profiling Reveals Diagnostic Models for Rheumatoid Arthritis
- PMID: 35514972
- PMCID: PMC9065411
- DOI: 10.3389/fimmu.2022.884462
Serum Antigenome Profiling Reveals Diagnostic Models for Rheumatoid Arthritis
Abstract
Objective: The study aimed to investigate the serum antigenomic profiling in rheumatoid arthritis (RA) and determine potential diagnostic biomarkers using label-free proteomic technology implemented with machine-learning algorithm.
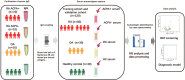
Method: Serum antigens were captured from a cohort consisting of 60 RA patients (45 ACPA-positive RA patients and 15 ACPA-negative RA patients), together with sex- and age-matched 30 osteoarthritis (OA) patients and 30 healthy controls. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was then performed. The significantly upregulated and downregulated proteins with fold change > 1.5 (p < 0.05) were selected. Based on these differentially expressed proteins (DEPs), a machine learning model was trained and validated to classify RA, ACPA-positive RA, and ACPA-negative RA.
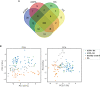
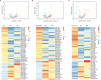
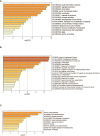
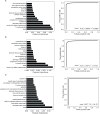
Results: We identified 62, 71, and 49 DEPs in RA, ACPA-positive RA, and ACPA-negative RA, respectively, as compared to OA and healthy controls. Typical pathway enrichment and protein-protein interaction networks were shown among these DEPs. Three panels were constructed to classify RA, ACPA-positive RA, and ACPA-negative RA using random forest models algorithm based on the molecular signature of DEPs, whose area under curve (AUC) were calculated as 0.9949 (95% CI = 0.9792-1), 0.9913 (95% CI = 0.9653-1), and 1.0 (95% CI = 1-1).
Conclusion: This study illustrated the serum auto-antigen profiling of RA. Among them, three panels of antigens were identified as diagnostic biomarkers to classify RA, ACPA-positive, and ACPA-negative RA patients.
Keywords: antigenome; biomarkers; mass spectrometry; random forest; rheumatoid arthritis.
Copyright © 2022 Han, Hou, Zheng, Cao, Shi, Zhang, Ye, Pan, Liu, Li, Hu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

