Can ERAP1 and ERAP2 Form Functional Heterodimers? A Structural Dynamics Investigation
- PMID: 35514997
- PMCID: PMC9065437
- DOI: 10.3389/fimmu.2022.863529
Can ERAP1 and ERAP2 Form Functional Heterodimers? A Structural Dynamics Investigation
Abstract
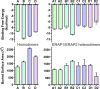
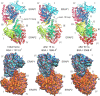
Endoplasmic reticulum aminopeptidases 1 and 2 (ERAP1 and ERAP2) play important roles in the generation of antigenic peptides presented by Major Histocompatibility Class I (MHCI) molecules and indirectly regulate adaptive immune responses. Although the discrete function of these enzymes has been extensively characterized, recent reports have suggested that they can also form heterodimers with functional consequences. However, lack of structural characterization of a putative ERAP1/ERAP2 dimer has limited our understanding of its biological role and significance. To address this, we employed computational molecular dynamics calculations to explore the topology of interactions between these two, based on experimentally determined homo-dimerization interfaces observed in crystal structures of ERAP2 or homologous enzymes. Our analysis of 8 possible dimerization models, suggested that the most likely ERAP1/ERAP2 heterodimerization topology involves the exon 10 loop, a non-conserved loop previously implicated in interactions between ERAP1 and the disulfide-bond shuffling chaperone ERp44. This dimerization topology allows access to the active site of both enzymes and is consistent with a previously reported construct in which ERAP1 and ERAP2 were linked by Fos/Jun zipper tags. The proposed model constitutes a tentative structural template to help understand the physiological role and significance of ERAP1/ERAP2 molecular interactions.
Keywords: aminopeptidase; MHC class I; adaptive immunity; antigen presentation; antigen processing; binding free energy; enzyme mechanism; molecular dynamics.
Copyright © 2022 Papakyriakou, Mpakali and Stratikos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
To Be or Not to Be: The Case of Endoplasmic Reticulum Aminopeptidase 2.Front Immunol. 2022 Jun 13;13:902567. doi: 10.3389/fimmu.2022.902567. eCollection 2022. Front Immunol. 2022. PMID: 35769458 Free PMC article. Review.
-
A systematic re-examination of processing of MHCI-bound antigenic peptide precursors by endoplasmic reticulum aminopeptidase 1.J Biol Chem. 2020 May 22;295(21):7193-7210. doi: 10.1074/jbc.RA120.012976. Epub 2020 Mar 17. J Biol Chem. 2020. PMID: 32184355 Free PMC article.
-
ERAP1-ERAP2 dimerization increases peptide-trimming efficiency.J Immunol. 2014 Jul 15;193(2):901-8. doi: 10.4049/jimmunol.1302855. Epub 2014 Jun 13. J Immunol. 2014. PMID: 24928998
-
Separate effects of the ankylosing spondylitis associated ERAP1 and ERAP2 aminopeptidases determine the influence of their combined phenotype on the HLA-B*27 peptidome.J Autoimmun. 2017 May;79:28-38. doi: 10.1016/j.jaut.2016.12.008. Epub 2017 Jan 4. J Autoimmun. 2017. PMID: 28063628
-
Regulation of ERAP1 and ERAP2 genes and their disfunction in human cancer.Hum Immunol. 2019 May;80(5):318-324. doi: 10.1016/j.humimm.2019.02.014. Epub 2019 Feb 27. Hum Immunol. 2019. PMID: 30825518 Review.
Cited by
-
Shedding Light on the Role of ERAP1 in Axial Spondyloarthritis.Cureus. 2023 Nov 14;15(11):e48806. doi: 10.7759/cureus.48806. eCollection 2023 Nov. Cureus. 2023. PMID: 38024089 Free PMC article. Review.
-
The ER Aminopeptidases, ERAP1 and ERAP2, synergize to self-modulate their respective activities.Front Immunol. 2022 Dec 8;13:1066483. doi: 10.3389/fimmu.2022.1066483. eCollection 2022. Front Immunol. 2022. PMID: 36569828 Free PMC article.
-
A Comparative Review of Pregnancy and Cancer and Their Association with Endoplasmic Reticulum Aminopeptidase 1 and 2.Int J Mol Sci. 2023 Feb 9;24(4):3454. doi: 10.3390/ijms24043454. Int J Mol Sci. 2023. PMID: 36834865 Free PMC article. Review.
-
To Be or Not to Be: The Case of Endoplasmic Reticulum Aminopeptidase 2.Front Immunol. 2022 Jun 13;13:902567. doi: 10.3389/fimmu.2022.902567. eCollection 2022. Front Immunol. 2022. PMID: 35769458 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

