ASPM Is a Prognostic Biomarker and Correlates With Immune Infiltration in Kidney Renal Clear Cell Carcinoma and Liver Hepatocellular Carcinoma
- PMID: 35515103
- PMCID: PMC9065448
- DOI: 10.3389/fonc.2022.632042
ASPM Is a Prognostic Biomarker and Correlates With Immune Infiltration in Kidney Renal Clear Cell Carcinoma and Liver Hepatocellular Carcinoma
Erratum in
-
Corrigendum: ASPM is a prognostic biomarker and correlates with immune infiltration in kidney renal clear cell carcinoma and liver hepatocellular carcinoma.Front Oncol. 2022 Aug 22;12:979968. doi: 10.3389/fonc.2022.979968. eCollection 2022. Front Oncol. 2022. PMID: 36072794 Free PMC article.
Abstract
Background: Abnormal spindle microtubule assembly (ASPM) is a centrosomal protein and that is related to a poor clinical prognosis and recurrence. However, the relationship between ASPM expression, tumor immunity, and the prognosis of different cancers remains unclear.
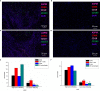
Methods: ASPM expression and its influence on tumor prognosis were analyzed using the Tumor Immune Estimation Resource (TIMER), UALCAN, OncoLnc, and Gene Expression Profiling Interactive Analysis (GEPIA) databases. The relationship between ASPM expression and tumor immunity was analyzed using the TIMER and GEPIA databases, and the results were further verified using qPCR, western blot, and multiplex quantitative immuno fluorescence.
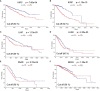
Results: The results showed that ASPM expression was significantly higher in most cancer tissues than in corresponding normal tissues, including kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), pancreatic adenocarcinoma (PAAD), and breast invasive carcinoma (BRCA). ASPM expression was significantly higher in late-stage cancers than in early-stages cancers (e.g., KIRC, KIRP, LIHC, LUAD, and BRCA; p < 0.05), demonstrating a possible role of ASPM in cancer progression and invasion. Moreover, our data showed that high ASPM expression was associated with poor overall survival, and disease-specific survival in KIRC and LIHC (p < 0.05). Besides, Cox hazard regression analysis results showed that ASPM may be an independent prognostic factor for KIRC and LIHC. ASPM expression showed a strong correlation with tumor-infiltrating B cells, CD8+ T cells, and M2 macrophages in KIRC and LIHC.
Conclusions: These findings demonstrate that the high expression of ASPM indicates poor prognosis as well as increased levels of immune cell infiltration in KIRC and LIHC. ASPM expression may serve as a novel prognostic biomarker for both the clinical outcome and immune cell infiltration in KIRC and LIHC.
Keywords: ASPM; biomarker; gene expression; prognosis; tumor-infiltrating.
Copyright © 2022 Deng, Liu, Zhuang, Tang and Huo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures.Front Immunol. 2021 Aug 31;12:721030. doi: 10.3389/fimmu.2021.721030. eCollection 2021. Front Immunol. 2021. PMID: 34531868 Free PMC article.
-
Prognostic Value of Long Noncoding RNA DLEU2 and Its Relationship with Immune Infiltration in Kidney Renal Clear Cell Carcinoma and Liver Hepatocellular Carcinoma.Int J Gen Med. 2021 Nov 11;14:8047-8064. doi: 10.2147/IJGM.S336428. eCollection 2021. Int J Gen Med. 2021. PMID: 34795513 Free PMC article.
-
Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers.Comput Struct Biotechnol J. 2022 Jun 18;20:3106-3119. doi: 10.1016/j.csbj.2022.06.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782736 Free PMC article.
-
The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism.Exp Mol Med. 2019 Jun 20;51(6):1-17. doi: 10.1038/s12276-019-0230-6. Exp Mol Med. 2019. PMID: 31221981 Free PMC article. Review.
-
Integrative data mining and meta-analysis to investigate the prognostic role of microRNA-200 family in various human malignant neoplasms: A consideration on heterogeneity.Gene. 2019 Oct 20;716:144025. doi: 10.1016/j.gene.2019.144025. Epub 2019 Aug 5. Gene. 2019. PMID: 31394177 Review.
Cited by
-
NCPAD2 is a favorable predictor of prognostic and immunotherapeutic biomarker for multiple cancer types including lung cancer.Genes Environ. 2024 Jan 3;46(1):2. doi: 10.1186/s41021-023-00291-4. Genes Environ. 2024. PMID: 38172945 Free PMC article.
-
FAM107A Inhibits the Growth, Invasion and Aerobic Glycolysis of LUAD Cells by Regulating CRYAB/PI3K/AKT.Biochem Genet. 2025 Jan 3. doi: 10.1007/s10528-024-11006-x. Online ahead of print. Biochem Genet. 2025. PMID: 39751722
-
Knockdown of TOP2A suppresses IL-17 signaling pathway and alleviates the progression of ulcerative colitis.Immun Inflamm Dis. 2024 Apr;12(4):e1207. doi: 10.1002/iid3.1207. Immun Inflamm Dis. 2024. PMID: 38661103 Free PMC article.
-
Omics Integration Uncovers Mechanisms Associated with HIV Viral Load and Potential Therapeutic Insights.medRxiv [Preprint]. 2025 Jul 30:2025.07.29.25332397. doi: 10.1101/2025.07.29.25332397. medRxiv. 2025. PMID: 40766151 Free PMC article. Preprint.
-
Unravelling infiltrating T-cell heterogeneity in kidney renal clear cell carcinoma: Integrative single-cell and spatial transcriptomic profiling.J Cell Mol Med. 2024 Jun;28(12):e18403. doi: 10.1111/jcmm.18403. J Cell Mol Med. 2024. PMID: 39031800 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

