Circular and linear: a tale of aptamer selection for the activation of SIRT1 to induce death in cancer cells
- PMID: 35516259
- PMCID: PMC9058605
- DOI: 10.1039/d0ra07857c
Circular and linear: a tale of aptamer selection for the activation of SIRT1 to induce death in cancer cells
Abstract
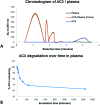
It is a challenge to select the right target to treat conditions without affecting non-diseased cells. Cancer belongs to the top 10 causes of death in the world and it remains difficult to treat. Amongst cancer emerging targets, silent information regulator 1 (SIRT1) - a histone deacetylase - has shown many roles in cancer, ageing and metabolism. Here we report novel SIRT1 ligands that bind and modulate the activity of SIRT1 within cells and enhance its enzymatic activity. We developed a modified aptamer capable of binding to and forming a complex with SIRT1. Our ligands are aptamers, they can be made of DNA or RNA oligonucleotides, their binding domain can recognise a target with very high affinity and specificity. We used the systematic evolution of ligands by exponential enrichment (SELEX) technique to develop circular and linear aptamers selectively binding to SIRT1. Cellular consequences of the interaction were monitored by fluorescence microscopy, cell viability assay, stability and enzymatic assays. Our results indicate that from our pool of aptamers, circular AC3 penetrates cancerous cells and is recruited to modulate the SIRT1 activity. This modulation of SIRT1 resulted in anticancer activity on different cancer cell lines. Furthermore, this modified aptamer showed no toxicity on one non-cancerous cell line and was stable in human plasma. We have demonstrated that aptamers are efficient tools for localisation of internal cell targets, and in this particular case, anticancer activity through modulation of SIRT1.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
The Effects of SELEX Conditions on the Resultant Aptamer Pools in the Selection of Aptamers Binding to Bacterial Cells.J Mol Evol. 2015 Dec;81(5-6):194-209. doi: 10.1007/s00239-015-9711-y. Epub 2015 Nov 4. J Mol Evol. 2015. PMID: 26538121
-
[Efficient screening for 8-oxoguanine DNA glycosylase binding aptamers via capillary electrophoresis].Se Pu. 2021 Jul 8;39(7):721-729. doi: 10.3724/SP.J.1123.2020.12017. Se Pu. 2021. PMID: 34227370 Free PMC article. Chinese.
-
An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes.Methods. 2016 Mar 15;97:51-7. doi: 10.1016/j.ymeth.2015.12.005. Epub 2015 Dec 8. Methods. 2016. PMID: 26678795
-
SELEX Modifications and Bioanalytical Techniques for Aptamer-Target Binding Characterization.Crit Rev Anal Chem. 2016 Nov;46(6):521-37. doi: 10.1080/10408347.2016.1157014. Epub 2016 Mar 15. Crit Rev Anal Chem. 2016. PMID: 26980177 Review.
-
Chimeric aptamers in cancer cell-targeted drug delivery.Crit Rev Biochem Mol Biol. 2011 Dec;46(6):459-77. doi: 10.3109/10409238.2011.614592. Epub 2011 Sep 28. Crit Rev Biochem Mol Biol. 2011. PMID: 21955150 Free PMC article. Review.
Cited by
-
The multifaceted roles of circular RNAs in cancer hallmarks: From mechanisms to clinical implications.Mol Ther Nucleic Acids. 2024 Jul 20;35(3):102286. doi: 10.1016/j.omtn.2024.102286. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39188305 Free PMC article. Review.
-
Recent Advances of Circular RNAs as Biomarkers for Osteosarcoma.Int J Gen Med. 2023 Jan 15;16:173-183. doi: 10.2147/IJGM.S380834. eCollection 2023. Int J Gen Med. 2023. PMID: 36687163 Free PMC article. Review.
References
-
- Olmos Y. Brosens J. J. Lam E. W. F. Drug Resist. Updates. 2011;14:35–44. - PubMed
LinkOut - more resources
Full Text Sources

