Interactome and Ubiquitinome Analyses Identify Functional Targets of Herpes Simplex Virus 1 Infected Cell Protein 0
- PMID: 35516420
- PMCID: PMC9062659
- DOI: 10.3389/fmicb.2022.856471
Interactome and Ubiquitinome Analyses Identify Functional Targets of Herpes Simplex Virus 1 Infected Cell Protein 0
Abstract
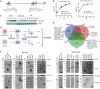
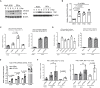
Herpes simplex virus 1 (HSV-1) can productively infect multiple cell types and establish latent infection in neurons. Infected cell protein 0 (ICP0) is an HSV-1 E3 ubiquitin ligase crucial for productive infection and reactivation from latency. However, our knowledge about its targets especially in neuronal cells is limited. We confirmed that, like in non-neuronal cells, ICP0-null virus exhibited major replication defects in primary mouse neurons and Neuro-2a cells. We identified many ICP0-interacting proteins in Neuro-2a cells, 293T cells, and human foreskin fibroblasts by mass spectrometry-based interactome analysis. Co-immunoprecipitation assays validated ICP0 interactions with acyl-coenzyme A thioesterase 8 (ACOT8), complement C1q binding protein (C1QBP), ovarian tumour domain-containing protein 4 (OTUD4), sorting nexin 9 (SNX9), and vimentin (VIM) in both Neuro-2a and 293T cells. Overexpression and knockdown experiments showed that SNX9 restricted replication of an ICP0-null but not wild-type virus in Neuro-2a cells. Ubiquitinome analysis by immunoprecipitating the trypsin-digested ubiquitin reminant followed by mass spectrometry identified numerous candidate ubiquitination substrates of ICP0 in infected Neuro-2a cells, among which OTUD4 and VIM were novel substrates confirmed to be ubiquitinated by transfected ICP0 in Neuro-2a cells despite no evidence of their degradation by ICP0. Expression of OTUD4 was induced independently of ICP0 during HSV-1 infection. Overexpressed OTUD4 enhanced type I interferon expression during infection with the ICP0-null but not wild-type virus. In summary, by combining two proteomic approaches followed by confirmatory and functional experiments, we identified and validated multiple novel targets of ICP0 and revealed potential restrictive activities of SNX9 and OTUD4 in neuronal cells.
Keywords: ICP0; herpes simplex virus; interactome; proteomics; ubiquitinome.
Copyright © 2022 Hou, Sun, Deng, Chen, Yang, Ji, Zhou, Ren and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
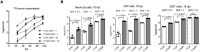



References
-
- Alandijany T., Roberts A. P. E., Conn K. L., Loney C., Mcfarlane S., Orr A., et al. (2018). Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 14:e1006769. 10.1371/journal.ppat.1006769 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

