Prediction and Inferred Evolution of Acid Tolerance Genes in the Biotechnologically Important Acidihalobacter Genus
- PMID: 35516430
- PMCID: PMC9062700
- DOI: 10.3389/fmicb.2022.848410
Prediction and Inferred Evolution of Acid Tolerance Genes in the Biotechnologically Important Acidihalobacter Genus
Abstract
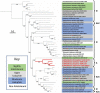
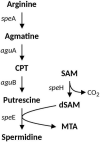
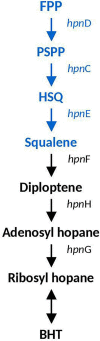
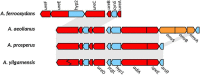
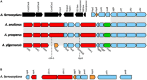
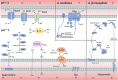
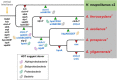
Acidihalobacter is a genus of acidophilic, gram-negative bacteria known for its ability to oxidize pyrite minerals in the presence of elevated chloride ions, a capability rare in other iron-sulfur oxidizing acidophiles. Previous research involving Acidihalobacter spp. has focused on their applicability in saline biomining operations and their genetic arsenal that allows them to cope with chloride, metal and oxidative stress. However, an understanding of the molecular adaptations that enable Acidihalobacter spp. to thrive under both acid and chloride stress is needed to provide a more comprehensive understanding of how this genus can thrive in such extreme biomining conditions. Currently, four genomes of the Acidihalobacter genus have been sequenced: Acidihalobacter prosperus DSM 5130T, Acidihalobacter yilgarnensis DSM 105917T, Acidihalobacter aeolianus DSM 14174T, and Acidihalobacter ferrooxydans DSM 14175T. Phylogenetic analysis shows that the Acidihalobacter genus roots to the Chromatiales class consisting of mostly halophilic microorganisms. In this study, we aim to advance our knowledge of the genetic repertoire of the Acidihalobacter genus that has enabled it to cope with acidic stress. We provide evidence of gene gain events that are hypothesized to help the Acidihalobacter genus cope with acid stress. Potential acid tolerance mechanisms that were found in the Acidihalobacter genomes include multiple potassium transporters, chloride/proton antiporters, glutamate decarboxylase system, arginine decarboxylase system, urease system, slp genes, squalene synthesis, and hopanoid synthesis. Some of these genes are hypothesized to have entered the Acidihalobacter via vertical decent from an inferred non-acidophilic ancestor, however, horizontal gene transfer (HGT) from other acidophilic lineages is probably responsible for the introduction of many acid resistance genes.
Keywords: acid resistance; chloride/proton antiporters; extreme acidophile; genome evolution; phylogenomics; polyextremophile; potassium transporters; urease.
Copyright © 2022 Boase, González, Vergara, Neira, Holmes and Watkin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov.Int J Syst Evol Microbiol. 2020 Dec;70(12):6226-6234. doi: 10.1099/ijsem.0.004519. Int J Syst Evol Microbiol. 2020. PMID: 33112221 Free PMC article.
-
Uncovering the Mechanisms of Halotolerance in the Extremely Acidophilic Members of the Acidihalobacter Genus Through Comparative Genome Analysis.Front Microbiol. 2019 Feb 8;10:155. doi: 10.3389/fmicb.2019.00155. eCollection 2019. Front Microbiol. 2019. PMID: 30853944 Free PMC article.
-
Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus.Genes (Basel). 2020 Nov 24;11(12):1392. doi: 10.3390/genes11121392. Genes (Basel). 2020. PMID: 33255299 Free PMC article.
-
In a quest for engineering acidophiles for biomining applications: challenges and opportunities.Genes (Basel). 2018 Feb 21;9(2):116. doi: 10.3390/genes9020116. Genes (Basel). 2018. PMID: 29466321 Free PMC article. Review.
-
Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments.Environ Microbiol. 2012 Jul;14(7):1597-611. doi: 10.1111/j.1462-2920.2011.02626.x. Epub 2011 Nov 3. Environ Microbiol. 2012. PMID: 22050575 Review.
Cited by
-
Process optimization and extraction of alkaline protease from halotolerant Streptomyces sp. VITGS3 and its use as a contact lens cleaner.J Genet Eng Biotechnol. 2025 Mar;23(1):100459. doi: 10.1016/j.jgeb.2025.100459. Epub 2025 Jan 18. J Genet Eng Biotechnol. 2025. PMID: 40074433 Free PMC article.
-
Extremophilic Microorganisms as a Source of Emerging Enzymes for the Food Industry: A Review.Food Sci Nutr. 2024 Dec 30;13(1):e4540. doi: 10.1002/fsn3.4540. eCollection 2025 Jan. Food Sci Nutr. 2024. PMID: 39803234 Free PMC article. Review.
-
Microbial adaptations to acidic, nutrient- and metal-rich lakes in Aotearoa New Zealand.Extremophiles. 2025 Jun 28;29(2):24. doi: 10.1007/s00792-025-01393-3. Extremophiles. 2025. PMID: 40580234 Free PMC article.
-
Understanding a Core Pilin of the Type IVa Pili of Acidithiobacillus thiooxidans, PilV.J Microbiol Biotechnol. 2024 Mar 28;34(3):527-537. doi: 10.4014/jmb.2310.10033. Epub 2024 Jan 17. J Microbiol Biotechnol. 2024. PMID: 38346803 Free PMC article.
-
Genomic insights into key mechanisms for carbon, nitrogen, and phosphate assimilation by the acidophilic, halotolerant genus Acidihalobacter members.FEMS Microbiol Ecol. 2024 Nov 23;100(12):fiae145. doi: 10.1093/femsec/fiae145. FEMS Microbiol Ecol. 2024. PMID: 39496518 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

