Metabolic characterisation of Magnetospirillum gryphiswaldense MSR-1 using LC-MS-based metabolite profiling
- PMID: 35516490
- PMCID: PMC9056635
- DOI: 10.1039/d0ra05326k
Metabolic characterisation of Magnetospirillum gryphiswaldense MSR-1 using LC-MS-based metabolite profiling
Abstract
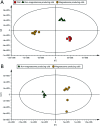
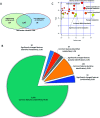
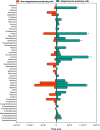
Magnetosomes are nano-sized magnetic nanoparticles with exquisite properties that can be used in a wide range of healthcare and biotechnological applications. They are biosynthesised by magnetotactic bacteria (MTB), such as Magnetospirillum gryphiswaldense MSR-1 (Mgryph). However, magnetosome bioprocessing yields low quantities compared to chemical synthesis of magnetic nanoparticles. Therefore, an understanding of the intracellular metabolites and metabolic networks related to Mgryph growth and magnetosome formation are vital to unlock the potential of this organism to develop improved bioprocesses. In this work, we investigated the metabolism of Mgryph using untargeted metabolomics. Liquid chromatography-mass spectrometry (LC-MS) was performed to profile spent medium samples of Mgryph cells grown under O2-limited (n = 6) and O2-rich conditions (n = 6) corresponding to magnetosome- and non-magnetosome producing cells, respectively. Multivariate, univariate and pathway enrichment analyses were conducted to identify significantly altered metabolites and pathways. Rigorous metabolite identification was carried out using authentic standards, the Mgryph-specific metabolite database and MS/MS mzCloud database. PCA and OPLS-DA showed clear separation and clustering of sample groups with cross-validation values of R2X = 0.76, R2Y = 0.99 and Q2 = 0.98 in OPLS-DA. As a result, 50 metabolites linked to 45 metabolic pathways were found to be significantly altered in the tested conditions, including: glycine, serine and threonine; butanoate; alanine, aspartate and glutamate metabolism; aminoacyl-tRNA biosynthesis and; pyruvate and citric acid cycle (TCA) metabolisms. Our findings demonstrate the potential of LC-MS to characterise key metabolites in Mgryph and will contribute to further understanding the metabolic mechanisms that affect Mgryph growth and magnetosome formation.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Wilhelm S. Tavares A. J. Dai Q. Ohta S. Audet J. Dvorak H. F. Chan W. C. W. Nat. Rev. Mater. 2016;1:16014. doi: 10.1038/natrevmats.2016.14. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

