Microscopy-BIDS: An Extension to the Brain Imaging Data Structure for Microscopy Data
- PMID: 35516811
- PMCID: PMC9063519
- DOI: 10.3389/fnins.2022.871228
Microscopy-BIDS: An Extension to the Brain Imaging Data Structure for Microscopy Data
Abstract
The Brain Imaging Data Structure (BIDS) is a specification for organizing, sharing, and archiving neuroimaging data and metadata in a reusable way. First developed for magnetic resonance imaging (MRI) datasets, the community-led specification evolved rapidly to include other modalities such as magnetoencephalography, positron emission tomography, and quantitative MRI (qMRI). In this work, we present an extension to BIDS for microscopy imaging data, along with example datasets. Microscopy-BIDS supports common imaging methods, including 2D/3D, ex/in vivo, micro-CT, and optical and electron microscopy. Microscopy-BIDS also includes comprehensible metadata definitions for hardware, image acquisition, and sample properties. This extension will facilitate future harmonization efforts in the context of multi-modal, multi-scale imaging such as the characterization of tissue microstructure with qMRI.
Keywords: data sharing; data structure; microscopy; open science; specification; standardization.
Copyright © 2022 Bourget, Kamentsky, Ghosh, Mazzamuto, Lazari, Markiewicz, Oostenveld, Niso, Halchenko, Lipp, Takerkart, Toussaint, Khan, Nilsonne, Castelli, The BIDS Maintainers and Cohen-Adad.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
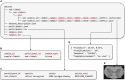
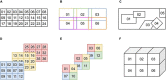
References
-
- Bandrowski A., Grethe J. S., Pilko A., Gillespie T., Pine G., Patel B., et al. (2021). SPARC Data Structure: rationale and Design of a FAIR Standard for Biomedical Research Data. bioRxiv [Preprint]. 10.1101/2021.02.10.430563 - DOI
-
- Benninger K., Hood G., Simmel D., Tuite L., Wetzel A., Ropelewski A., et al. (2020). “Cyberinfrastructure of a multi-petabyte microscopy resource for neuroscience research,” in Proceedings of the Practice and Experience in Advanced Research Computing (PEARC ’20) (New York, NY: Association for Computing Machinery; ), 1–7. 10.1145/3311790.3396653 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

