Investigation of some benzoquinazoline and quinazoline derivatives as novel inhibitors of HCV-NS3/4A protease: biological, molecular docking and QSAR studies
- PMID: 35517076
- PMCID: PMC9056986
- DOI: 10.1039/d0ra05604a
Investigation of some benzoquinazoline and quinazoline derivatives as novel inhibitors of HCV-NS3/4A protease: biological, molecular docking and QSAR studies
Abstract
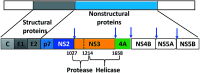
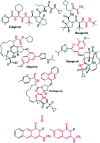
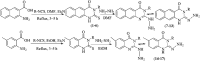
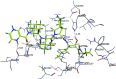
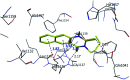
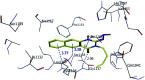
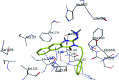
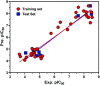
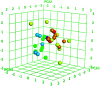
Morbidity and mortality due to hepatitis C virus (HCV) is a globe health concern. Hence, there is a persistent demand to design and optimize current HCV therapy and develop novel agents. HCV NS3/A4 protease plays an essential role in HCV life cycle and replication. Thus, HCV NS3/A4 protease inhibitors are one of the best therapeutic targets for the identification of novel candidate drugs. Recent studies have shown some benzoquinazolines as potent antiviral agents and promising HAV-3C protease inhibitors. In the present study, a series of benzo[g]quinazolines (1-13) and their quinazoline analogues (14-17) were evaluated for their HCV-NS3/4A inhibitory activities using in vitro assay. Our results revealed that the target compounds inhibited the activity of the NS3/4A enzyme, (IC50 = 6.41 ± 0.12 to 78.80 ± 1.70 μM) in comparison to telaprevir (IC50 = 1.72 ± 0.03 μM) as a reference drug. Compounds 1, 2, 3, 9, 10 and 13 showed the highest activity (IC50 = 11.02 ± 0.25, 6.41 ± 0.12, 9.35 ± 0.19, 9.08 ± 0.20, 16.03 ± 0.34 and 7.21 ± 0.15 μM, respectively). Molecular docking was performed to study the binding modes of the docked-chosen benzo[g]quinazolines, hydrogen bonding, and amino acid residues at the catalytic triad of the NS3/4A enzyme of HCV. The QSAR was determined to explore the relationships between the molecular structures of the targets and their biological activities by developing prediction models among the known HCV NS3/A4 inhibitors and then to predict the inhibitory activity of the target molecules synthesized.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

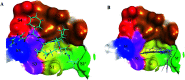
Similar articles
-
Identification of novel small molecule inhibitors against the NS3/4A protease of hepatitis C virus genotype 4a.Curr Pharm Des. 2018;24(37):4484-4491. doi: 10.2174/1381612825666181203153835. Curr Pharm Des. 2018. PMID: 30501598
-
A Novel Approach to Develop New and Potent Inhibitors for the Simultaneous Inhibition of Protease and Helicase Activities of HCV NS3/4A Protease: A Computational Approach.Molecules. 2023 Jan 29;28(3):1300. doi: 10.3390/molecules28031300. Molecules. 2023. PMID: 36770965 Free PMC article.
-
Discovery and development of VX-950, a novel, covalent, and reversible inhibitor of hepatitis C virus NS3.4A serine protease.Infect Disord Drug Targets. 2006 Mar;6(1):3-16. doi: 10.2174/187152606776056706. Infect Disord Drug Targets. 2006. PMID: 16787300 Review.
-
Computer aided screening of Accacia nilotica phytochemicals against HCV NS3/4a.Bioinformation. 2013 Aug 7;9(14):710-4. doi: 10.6026/97320630009710. eCollection 2013. Bioinformation. 2013. PMID: 23976825 Free PMC article.
-
Discovery of small-molecule inhibitors of HCV NS3-4A protease as potential therapeutic agents against HCV infection.Curr Med Chem. 2005;12(20):2317-42. doi: 10.2174/0929867054864769. Curr Med Chem. 2005. PMID: 16181135 Review.
Cited by
-
Therapeutic Intervention of Serine Protease Inhibitors against Hepatitis C Virus.Curr Med Chem. 2024;31(15):2052-2072. doi: 10.2174/0109298673234823230921090431. Curr Med Chem. 2024. PMID: 37855348
-
Antiproliferative and Antiangiogenic Properties of New VEGFR-2-targeting 2-thioxobenzo[g]quinazoline Derivatives (In Vitro).Molecules. 2020 Dec 15;25(24):5944. doi: 10.3390/molecules25245944. Molecules. 2020. PMID: 33333992 Free PMC article.
-
Benzo[g]quinazolines as antifungal against candidiasis: Screening, molecular docking, and QSAR investigations.Saudi Pharm J. 2023 Jun;31(6):815-823. doi: 10.1016/j.jsps.2023.04.012. Epub 2023 Apr 15. Saudi Pharm J. 2023. PMID: 37228321 Free PMC article.
-
Evaluation of Some Benzo[g]Quinazoline Derivatives as Antiviral Agents against Human Rotavirus Wa Strain: Biological Screening and Docking Study.Curr Issues Mol Biol. 2023 Mar 14;45(3):2409-2421. doi: 10.3390/cimb45030156. Curr Issues Mol Biol. 2023. PMID: 36975526 Free PMC article.
-
Significant pharmacological activities of benzoquinazolines scaffold.Pharmacol Rep. 2023 Apr;75(2):223-235. doi: 10.1007/s43440-023-00453-9. Epub 2023 Feb 6. Pharmacol Rep. 2023. PMID: 36740656 Review.
References
LinkOut - more resources
Full Text Sources

