A sandwich-type bacteriophage-based amperometric biosensor for the detection of Shiga toxin-producing Escherichia coli serogroups in complex matrices
- PMID: 35517084
- PMCID: PMC9056931
- DOI: 10.1039/d0ra06223e
A sandwich-type bacteriophage-based amperometric biosensor for the detection of Shiga toxin-producing Escherichia coli serogroups in complex matrices
Abstract
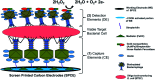
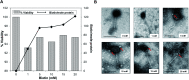
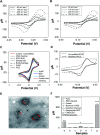
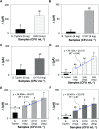
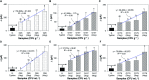
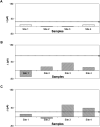
Immuno-based biosensors are a popular tool designed for pathogen screening and detection. The current antibody-based biosensors employ direct, indirect, or sandwich detection approaches; however, instability, cross-reactivity, and high-cost render them unreliable and impractical. To circumvent these drawbacks, here we report a portable sandwich-type bacteriophage-based amperometric biosensor, which is highly-specific to various Shiga toxin-producing Escherichia coli (STEC) serogroups. Environmentally isolated and biotinylated bacteriophages were directly immobilized onto a streptavidin-coated screen-printed carbon electrode (SPCE), which recognized and captured viable target cells. Samples (50 μL) were transferred to these bacteriophage-functionalized SPCEs (12 min, room temp) before sequentially adding a bacteriophage-gold nanoparticle solution (20 μL), H2O2 (40 mM), and 1,1'-ferrocenedicarboxylic acid for amperometric tests (100 mV s-1) and analysis (ANOVA and LSD, P < 0.05). The optimum biotin concentration (10 mM) retained 94.47% bacteriophage viability. Non-target bacteria (Listeria monocytogenes and Salmonella Typhimurium) had delta currents below the threshold of a positive detection. With less than 1 h turn-around time, the amperometric biosensor had a detection limit of 10-102 CFU mL-1 for STEC O157, O26, and O179 strains and R 2 values of 0.97, 0.99, and 0.87, respectively, and a similar detection limit was observed in complex matrices, 10-102 CFU g-1 or mL-1 with R 2 values of 0.98, 0.95, and 0.76, respectively. The newly developed portable amperometric biosensor was able to rapidly detect viable target cells at low inoculum levels, thus providing an inexpensive and improved alternative to the current immuno- and laboratory-based STEC screening methods.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
References
-
- Beutin L. Fach P. Microbiol. Spectrum. 2014;2:3. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

