Investigation of the target-site resistance of EPSP synthase mutants P106T and T102I/P106S against glyphosate
- PMID: 35517162
- PMCID: PMC9058485
- DOI: 10.1039/d0ra09061a
Investigation of the target-site resistance of EPSP synthase mutants P106T and T102I/P106S against glyphosate
Abstract
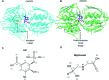
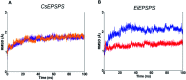
The shikimate pathway enzyme 5-enolpyruvyl shikimate-3-phosphate synthase (EPSPS) catalyzes the reaction involved in the production of amino acids essential for plant growth and survival. Thus, EPSPS is the main target of various herbicides, including glyphosate, a broad-spectrum herbicide that acts as a competitive inhibitor of phosphoenolpyruvate (PEP), which is the natural substrate of EPSPS. However, punctual mutations in the EPSPS gene have led to glyphosate resistance in some plants. Here, we investigated the mechanism of EPSPS resistance to glyphosate in mutants of two weed species, Conyza sumatrensis (mutant, P106T) and Eleusine indica (mutant, T102I/P106S), both of which have an economic impact on industrial crops. Molecular dynamics (MD) simulations and binding free energy calculations revealed the influence of the mutations on the affinity of glyphosate in the PEP-binding site. The amino acid residues of the EPSPS protein in both species involved in glyphosate resistance were elucidated as well as other residues that could be useful for protein engineering. In addition, during MD simulations, we identified conformational changes in glyphosate when complexed with resistant EPSPS, related to loss of herbicide activity and binding affinity. Our computational findings are consistent with previous experimental results and clarify the inhibitory activity of glyphosate as well as the structural target-site resistance of EPSPS against glyphosate.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
-
- Duke S. O. Powles S. B. Pest Manage. Sci. 2008;64:319–325. - PubMed
-
- Duke S. O. Pest Manage. Sci. 2018;74:1027–1034. - PubMed
-
- Maeda H. Dudareva N. Annu. Rev. Plant Biol. 2012;63:73–105. - PubMed
-
- Duke S. O. J. Agric. Food Chem. 2011;59:5835–5841. - PubMed
-
- dos Santos A. M. Lima A. H. Alves C. N. Lameira J. J. Phys. Chem. B. 2017;121:8626–8637. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

