Novel quercetin and apigenin-acetamide derivatives: design, synthesis, characterization, biological evaluation and molecular docking studies
- PMID: 35517443
- PMCID: PMC9055277
- DOI: 10.1039/d0ra04559d
Novel quercetin and apigenin-acetamide derivatives: design, synthesis, characterization, biological evaluation and molecular docking studies
Abstract
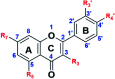
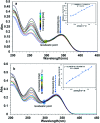
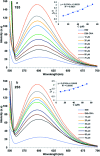
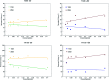
Flavonoids exhibit essential but limited biological properties which can be enhanced through chemical modifications. In this study, we designed, synthesized, and characterized two novel flavonoid derivatives, quercetin penta-acetamide (1S3) and apigenin tri-acetamide (2S3). These compounds were confirmed using (1H, 13C) NMR, UV-Vis, and FT-IR characterizations. Their interaction with fish sperm DNA (FS-DNA) at physiological pH was investigated by UV-Vis and fluorescence spectrophotometry. The binding constant (K b) for the UV-Vis experiment was found to be 1.43 ± 0.3 × 104 M-1 for 1S3 and 2.08 ± 0.2 × 104 M-1 for 2S3. The binding constants (K SV) for the fluorescence quenching experiment were 1.83 × 104 M-1 and 1.96 × 104 M-1 for 1S3 and 2S3, respectively. Based on molecular modeling and docking studies, the binding affinities were found to be -7.9 and -9.1 kcal mol-1, for 1S3 and 2S3, respectively. The compound-DNA docked model correlated with our experimental results, and they are groove binders. Furthermore, mutagenicity potential was examined. 1S3 and its metabolites showed no mutagenic activity for both TA98 and TA100 strains. 2S3 did not show any mutagenic activity for the strain TA 98, while its metabolites were only active at high doses. Both 2S3 and its metabolites showed mutagenic activity in the TA100 strain.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Mutagenicities of 61 flavonoids and 11 related compounds.Environ Mutagen. 1981;3(4):401-19. doi: 10.1002/em.2860030402. Environ Mutagen. 1981. PMID: 7021146
-
Mechanism of Interactions of dsDNA Binding with Apigenin and Its Sulfamate Derivatives Using Multispectroscopic, Voltammetric, and Molecular Docking Studies.ACS Omega. 2021 Feb 19;6(8):5124-5137. doi: 10.1021/acsomega.0c02612. eCollection 2021 Mar 2. ACS Omega. 2021. PMID: 33681554 Free PMC article.
-
Evaluation of DNA Binding, Radicals Scavenging and Antimicrobial Studies of Newly Synthesized N-Substituted Naphthalimides: Spectroscopic and Molecular Docking Investigations.J Fluoresc. 2015 Nov;25(6):1905-20. doi: 10.1007/s10895-015-1683-1. Epub 2015 Oct 13. J Fluoresc. 2015. PMID: 26462815
-
Isolation and studies of the mutagenic activity in the Ames test of flavonoids naturally occurring in medical herbs.Mutat Res. 1990 Mar;240(3):209-16. doi: 10.1016/0165-1218(90)90060-f. Mutat Res. 1990. PMID: 2179716
-
Synthesis, characterization and biological evaluation of novel benzimidazole derivatives.J Biomol Struct Dyn. 2020 Apr;38(6):1670-1682. doi: 10.1080/07391102.2019.1617783. Epub 2019 May 22. J Biomol Struct Dyn. 2020. PMID: 31074356
Cited by
-
Pharmacology and mechanisms of apigenin in preventing osteoporosis.Front Pharmacol. 2024 Dec 12;15:1486646. doi: 10.3389/fphar.2024.1486646. eCollection 2024. Front Pharmacol. 2024. PMID: 39726788 Free PMC article. Review.
-
Current overview of the mechanistic pathways and influence of physicochemical parameters on the microbial synthesis and applications of metallic nanoparticles.Bioprocess Biosyst Eng. 2025 Jun 25. doi: 10.1007/s00449-025-03190-w. Online ahead of print. Bioprocess Biosyst Eng. 2025. PMID: 40562943 Review.
-
Selective Structural Derivatization of Flavonoid Acetamides Significantly Impacts Their Bioavailability and Antioxidant Properties.Molecules. 2022 Nov 22;27(23):8133. doi: 10.3390/molecules27238133. Molecules. 2022. PMID: 36500226 Free PMC article.
-
Synthesis, biological and computational studies of flavonoid acetamide derivatives.RSC Adv. 2022 Mar 30;12(16):10037-10050. doi: 10.1039/d2ra01375d. eCollection 2022 Mar 25. RSC Adv. 2022. PMID: 35424949 Free PMC article.
-
Fisetin decreases the duration of ictal-like discharges in mouse hippocampal slices.J Biol Phys. 2022 Sep;48(3):355-368. doi: 10.1007/s10867-022-09612-0. Epub 2022 Aug 10. J Biol Phys. 2022. PMID: 35948819 Free PMC article.
References
-
- De Conti Lourenço R. M., Da Silva Melo P. and De Almeida A. B. A., in Antifungal Metabolites from Plants, Springer-Verlag, Berlin Heidelberg, 2013, pp. 283–300
LinkOut - more resources
Full Text Sources

