Variable and Conserved Regions of Secondary Structure in the β-Trefoil Fold: Structure Versus Function
- PMID: 35517858
- PMCID: PMC9062101
- DOI: 10.3389/fmolb.2022.889943
Variable and Conserved Regions of Secondary Structure in the β-Trefoil Fold: Structure Versus Function
Abstract
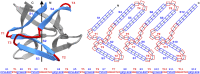
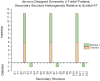
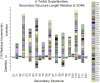
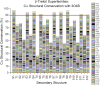
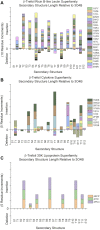
β-trefoil proteins exhibit an approximate C3 rotational symmetry. An analysis of the secondary structure for members of this diverse superfamily of proteins indicates that it is comprised of remarkably conserved β-strands and highly-divergent turn regions. A fundamental "minimal" architecture can be identified that is devoid of heterogenous and extended turn regions, and is conserved among all family members. Conversely, the different functional families of β-trefoils can potentially be identified by their unique turn patterns (or turn "signature"). Such analyses provide clues as to the evolution of the β-trefoil family, suggesting a folding/stability role for the β-strands and a functional role for turn regions. This viewpoint can also guide de novo protein design of β-trefoil proteins having novel functionality.
Keywords: de novo design; folding nucleus; hydrophobic patterning; ligand; protein symmetry.
Copyright © 2022 Blaber.
Conflict of interest statement
MB is a cofounder and has equity ownership in Trefoil Therapeutics Inc.
Figures

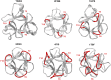

Similar articles
-
Conserved buried water molecules enable the β-trefoil architecture.Protein Sci. 2020 Aug;29(8):1794-1802. doi: 10.1002/pro.3899. Epub 2020 Jul 8. Protein Sci. 2020. PMID: 32542709 Free PMC article.
-
Ab initio folding of a trefoil-fold motif reveals structural similarity with a β-propeller blade motif.Protein Sci. 2020 May;29(5):1172-1185. doi: 10.1002/pro.3850. Epub 2020 Mar 25. Protein Sci. 2020. PMID: 32142181 Free PMC article.
-
Evidence for the emergence of β-trefoils by 'Peptide Budding' from an IgG-like β-sandwich.PLoS Comput Biol. 2022 Feb 14;18(2):e1009833. doi: 10.1371/journal.pcbi.1009833. eCollection 2022 Feb. PLoS Comput Biol. 2022. PMID: 35157697 Free PMC article.
-
Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a β-trefoil fold.Biosci Biotechnol Biochem. 2013;77(7):1363-71. doi: 10.1271/bbb.130183. Epub 2013 Jul 7. Biosci Biotechnol Biochem. 2013. PMID: 23832347 Review.
-
Understanding the mechanism of beta-sheet folding from a chemical and biological perspective.Biopolymers. 2008;90(6):751-8. doi: 10.1002/bip.21101. Biopolymers. 2008. PMID: 18844292 Review.
Cited by
-
Engineering the kinetic stability of a β-trefoil protein by tuning its topological complexity.Front Mol Biosci. 2023 Feb 8;10:1021733. doi: 10.3389/fmolb.2023.1021733. eCollection 2023. Front Mol Biosci. 2023. PMID: 36845544 Free PMC article.
-
Canonical or noncanonical? Structural plasticity of serine protease-binding loops in Kunitz-STI protease inhibitors.Protein Sci. 2023 Feb;32(2):e4570. doi: 10.1002/pro.4570. Protein Sci. 2023. PMID: 36660780 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

