RNA-Binding Macrocyclic Peptides
- PMID: 35517859
- PMCID: PMC9062085
- DOI: 10.3389/fmolb.2022.883060
RNA-Binding Macrocyclic Peptides
Abstract
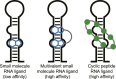
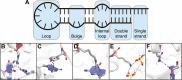
Being able to effectively target RNA with potent ligands will open up a large number of potential therapeutic options. The knowledge on how to achieve this is ever expanding but an important question that remains open is what chemical matter is suitable to achieve this goal. The high flexibility of an RNA as well as its more limited chemical diversity and featureless binding sites can be difficult to target selectively but can be addressed by well-designed cyclic peptides. In this review we will provide an overview of reported cyclic peptide ligands for therapeutically relevant RNA targets and discuss the methods used to discover them. We will also provide critical insights into the properties required for potent and selective interaction and suggestions on how to assess these parameters. The use of cyclic peptides to target RNA is still in its infancy but the lessons learned from past examples can be adopted for the development of novel potent and selective ligands.
Keywords: RNA binding; macrocyclic peptides; natural products; peptide library screening; structure-based design.
Copyright © 2022 Pal and ‘t Hart.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




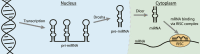






References
-
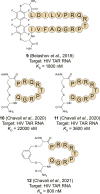
- Athanassiou Z., Dias R. L. A., Moehle K., Dobson N., Varani G., Robinson J. A. (2004). Structural Mimicry of Retroviral Tat Proteins by Constrained β-Hairpin Peptidomimetics: Ligands with High Affinity and Selectivity for Viral TAR RNA Regulatory Elements. J. Am. Chem. Soc. 126, 6906–6913. 10.1021/ja0497680 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

