Global and Comparative Proteome Signatures in the Lens Capsule, Trabecular Meshwork, and Iris of Patients With Pseudoexfoliation Glaucoma
- PMID: 35517867
- PMCID: PMC9065473
- DOI: 10.3389/fmolb.2022.877250
Global and Comparative Proteome Signatures in the Lens Capsule, Trabecular Meshwork, and Iris of Patients With Pseudoexfoliation Glaucoma
Abstract
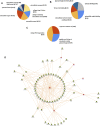
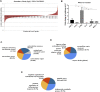
Pseudoexfoliation (PXF) is characterized by the accumulation of the exfoliative material in the eye and high rates of blindness if left untreated. Pseudoexfoliation glaucoma (PXG) is generally diagnosed too late due to its asymptomatic nature, necessitating the development of new effective screening tools for the early diagnosis of the disease. Thus, the increasing prevalence of this disease due to an aging population has demanded the identification of suitable biomarkers for the early detection of the disease or detection of the onset of glaucoma in the eyes with PXF. We applied a proteomics strategy based on a high-throughput screening method for the determination of proteins involving PXF and PXG pathogenesis. The lens capsule (LC), iris, and trabecular meshwork (TM) samples with PXF and PXG were taken by surgical trabeculectomy, and control samples were taken from the donor corneal buttons obtained from the institutional eye bank to characterize the proteome profile. Peptides from the LC were analyzed using liquid chromatography with tandem mass spectrometry (LC-MS/MS). The protein of interest and cytokine/chemokine profiles were verified using immunohistochemistry and the bio-plex kit assay, respectively. There were a total of 1433 proteins identified in the human LC, of which 27 proteins were overexpressed and eight proteins were underexpressed in PXG compared with PXF. Overexpressed proteins such as fibromodulin, decorin, lysyl oxidase homolog 1, collagen alpha-1(I) chain, collagen alpha-3(VI) chain, and biglycan were the major components of the extracellular matrix (ECM) proteins involved in cell-matrix interactions or ECM proteoglycans and the assembly and cross-linking of collagen fibrils. The ECM composition and homeostasis are altered in glaucoma. Thus, quantitative proteomics is a method to discover molecular markers in the eye. Monitoring these events can help evaluate disease progression in future studies.
Keywords: extracellular matrix; iris; lens capsule; lysyl oxidase; mass spectroscopy; pseudoexfoliation; pseudoexfoliation glaucoma; trabecular meshwork.
Copyright © 2022 Sahay, Chakraborty and Rao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Anderson D. R., Hendrickson A. (1974). Effect of Intraocular Pressure on Rapid Axoplasmic Transport in Monkey Optic Nerve. Invest. Ophthalmol. 13, 771–783. - PubMed
-
- Babizhayev M. A. (2012). Biomarkers and Special Features of Oxidative Stress in the Anterior Segment of the Eye Linked to Lens Cataract and the Trabecular Meshwork Injury in Primary Open-Angle Glaucoma: Challenges of Dual Combination Therapy with N-Acetylcarnosine Lubricant Eye D. Fundam. Clin. Pharmacol. 26, 86–117. 10.1111/j.1472-8206.2011.00969.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

