A coumarin derivative-Cu2+ complex-based fluorescent chemosensor for detection of biothiols
- PMID: 35517943
- PMCID: PMC9057049
- DOI: 10.1039/d0ra05651k
A coumarin derivative-Cu2+ complex-based fluorescent chemosensor for detection of biothiols
Abstract
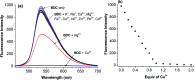
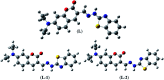
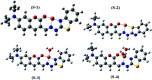
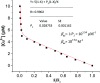
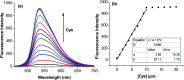
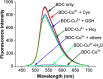
Herein, a novel fluorescent sensor has been developed for the detection of biothiols based on theoretical calculations of the stability constant of the complex between a Cu2+ ion and (E)-3-((2-(benzo[d]thiazol-2-yl)hydrazono)methyl)-7-(diethylamino) coumarin (BDC) as a fluorescent ligand. In this study, on the basis of density functional theory method, the Gibbs free energy of ligand-exchange reaction and the solvation model were carried out using thermodynamic cycles. The obtained results are in good agreement with the experimental data. The BDC-Cu2+ complex can be used as a fluorescent sensor for the detection of biothiols in the presence of non-thiol containing amino acids, with a detection limit for cysteine at 0.3 μM. Moreover, theoretical calculations of excited states were used to elucidate variations in the fluorescence properties. The computed results show that the excited doublet states D2 and D1 are dark doublet states, which quench the fluorescence of the complex.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
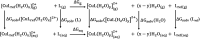
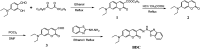

Similar articles
-
A simple fluorescent probe for the fast sequential detection of copper and biothiols based on a benzothiazole derivative.Spectrochim Acta A Mol Biomol Spectrosc. 2018 Feb 15;191:427-434. doi: 10.1016/j.saa.2017.09.069. Epub 2017 Sep 28. Spectrochim Acta A Mol Biomol Spectrosc. 2018. PMID: 29073543
-
A novel fluorescent chemosensor based on coumarin and quinolinyl-benzothiazole for sequential recognition of Cu2+ and PPi and its applicability in live cell imaging.Spectrochim Acta A Mol Biomol Spectrosc. 2020 Apr 5;230:118022. doi: 10.1016/j.saa.2019.118022. Epub 2020 Jan 3. Spectrochim Acta A Mol Biomol Spectrosc. 2020. PMID: 31927510
-
Dual chemosensor for the rapid detection of mercury(ii) pollution and biothiols.Analyst. 2019 Aug 21;144(16):4908-4916. doi: 10.1039/c9an01055f. Epub 2019 Jul 17. Analyst. 2019. PMID: 31312834
-
A Benzothiazole-Based Fluorescence Turn-on Sensor for Copper(II).J Fluoresc. 2021 Jul;31(4):1203-1209. doi: 10.1007/s10895-021-02752-x. Epub 2021 May 26. J Fluoresc. 2021. PMID: 34037894
-
A Copper (II) Ensemble-Based Fluorescence Chemosensor and Its Application in the 'Naked-Eye' Detection of Biothiols in Human Urine.Sensors (Basel). 2020 Feb 29;20(5):1331. doi: 10.3390/s20051331. Sensors (Basel). 2020. PMID: 32121408 Free PMC article.
Cited by
-
A Novel Fluorescent Sensor for Detecting Ag+ and Hg2+ ions: A Combination of Theoretical and Experimental Studies.J Fluoresc. 2024 Oct 23. doi: 10.1007/s10895-024-03988-z. Online ahead of print. J Fluoresc. 2024. PMID: 39441258
-
Trending Topics on Coumarin and Its Derivatives in 2020.Molecules. 2021 Jan 19;26(2):501. doi: 10.3390/molecules26020501. Molecules. 2021. PMID: 33477785 Free PMC article. Review.
-
A mercury complex-based fluorescent sensor for biological thiols.RSC Adv. 2025 Jun 13;15(25):20125-20133. doi: 10.1039/d5ra02268a. eCollection 2025 Jun 10. RSC Adv. 2025. PMID: 40519683 Free PMC article.
References
LinkOut - more resources
Full Text Sources

