G-Protein coupled receptors: structure and function in drug discovery
- PMID: 35517958
- PMCID: PMC9057076
- DOI: 10.1039/d0ra08003a
G-Protein coupled receptors: structure and function in drug discovery
Abstract
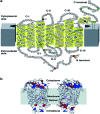
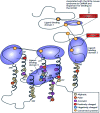
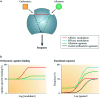
The G-protein coupled receptors (GPCRs) superfamily comprise similar proteins arranged into families or classes thus making it one of the largest in the mammalian genome. GPCRs take part in many vital physiological functions making them targets for numerous novel drugs. GPCRs share some distinctive features, such as the seven transmembrane domains, they also differ in the number of conserved residues in their transmembrane domain. Here we provide an introductory and accessible review detailing the computational advances in GPCR pharmacology and drug discovery. An overview is provided on family A-C GPCRs; their structural differences, GPCR signalling, allosteric binding and cooperativity. The dielectric constant (relative permittivity) of proteins is also discussed in the context of site-specific environmental effects.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

