Production of high-complexity frameshift neoantigen peptide microarrays
- PMID: 35518269
- PMCID: PMC9056171
- DOI: 10.1039/d0ra05267a
Production of high-complexity frameshift neoantigen peptide microarrays
Abstract
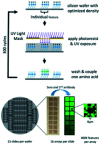
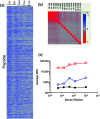
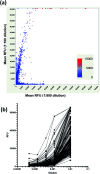
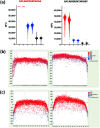
Parallel measurement of large numbers of antigen-antibody interactions are increasingly enabled by peptide microarray technologies. Our group has developed an in situ synthesized peptide microarray of >400 000 frameshift neoantigens using mask-based photolithographic peptide synthesis, to profile patient specific neoantigen reactive antibodies in a single assay. The system produces 208 replicate mircoarrays per wafer and is capable of producing multiple wafers per synthetic lot to routinely synthesize over 300 million peptides simultaneously. In this report, we demonstrate the feasibility of the system for detecting peripheral-blood antibody binding to frameshift neoantigens across multiple synthetic lots.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
S. A. J. and L. S. hold patents and pending patents with respect to frameshift antigens and FSP arrays. S. A. J. is a founder of Calviri which is commercializing these diagnostic arrays. K. S. and J. R. B. are employees of Calviri. All authors have ownership interests in Calviri.
Figures
Similar articles
-
Comparison of personal and shared frameshift neoantigen vaccines in a mouse mammary cancer model.BMC Immunol. 2020 May 5;21(1):25. doi: 10.1186/s12865-020-00350-3. BMC Immunol. 2020. PMID: 32370785 Free PMC article.
-
Neoantigen-reactive T cells exhibit effective anti-tumor activity against colorectal cancer.Hum Vaccin Immunother. 2022 Dec 31;18(1):1-11. doi: 10.1080/21645515.2021.1891814. Epub 2021 Mar 9. Hum Vaccin Immunother. 2022. PMID: 33689574 Free PMC article.
-
Neoantigen identification strategies enable personalized immunotherapy in refractory solid tumors.J Clin Invest. 2019 Mar 5;129(5):2056-2070. doi: 10.1172/JCI99538. Print 2019 May 1. J Clin Invest. 2019. PMID: 30835255 Free PMC article.
-
Beyond Sequencing: Prioritizing and Delivering Neoantigens for Cancer Vaccines.Methods Mol Biol. 2022;2410:649-670. doi: 10.1007/978-1-0716-1884-4_35. Methods Mol Biol. 2022. PMID: 34914074 Review.
-
Neoantigen vaccine platforms in clinical development: understanding the future of personalized immunotherapy.Expert Opin Investig Drugs. 2021 May;30(5):529-541. doi: 10.1080/13543784.2021.1896702. Epub 2021 Mar 31. Expert Opin Investig Drugs. 2021. PMID: 33641576 Free PMC article. Review.
Cited by
-
Predicting response and toxicity to immune checkpoint inhibitors in lung cancer using antibodies to frameshift neoantigens.J Transl Med. 2023 May 22;21(1):338. doi: 10.1186/s12967-023-04172-w. J Transl Med. 2023. PMID: 37217961 Free PMC article.
-
Vaccines for immunoprevention of DNA mismatch repair deficient cancers.J Immunother Cancer. 2022 Jun;10(6):e004416. doi: 10.1136/jitc-2021-004416. J Immunother Cancer. 2022. PMID: 35732349 Free PMC article. Review.
References
-
- Mattes D. S. Rentschler S. Foertsch T. C. Münch S. W. Loeffler F. F. Nesterov-Mueller A. Bräse S. Breitling F. Small Methods. 2018;2:1700205. doi: 10.1002/smtd.201700205. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

