Characterization and molecular docking of new Δ17 fatty acid desaturase genes from Rhizophagus irregularis and Octopus bimaculoides
- PMID: 35518462
- PMCID: PMC9061052
- DOI: 10.1039/c9ra00535h
Characterization and molecular docking of new Δ17 fatty acid desaturase genes from Rhizophagus irregularis and Octopus bimaculoides
Abstract
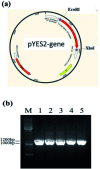
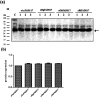
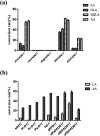
Fatty acid desaturases are key enzymes in the biosynthesis of n-3 polyunsaturated fatty acids (PUFAs) via conversion of n-6 polyunsaturates to their n-3 counterparts. In this study, we reported the characterization and molecular docking of Δ17 desaturases from Rhizophagus irregularis and Octopus bimaculoides. These two new desaturase genes were screened using the known Δ17 desaturase gene (oPaFADS17) from Pythium aphanidermatum as a template. Analysis of their genes revealed that the sequences of oRiFADS17 and oObFADS17 contained the typical His-rich motifs (one HXXXH and two HXXHH). They were then expressed in Saccharomyces cerevisiae INVSc1 to examine their activities and substrate preferences. Our results show that the two candidate n-3 desaturases possess a strong Δ17 desaturase activity, exhibiting remarkable increase in desaturation activity on C20 fatty acids compared to C18 fatty acids. To the best of our knowledge, oRiFADS17 desaturase has greater (3-4 fold) catalytic activity for C18 substrates than other reported Δ17 desaturases and oObFADS17 is the first reported Δ17 desaturase in sea mollusks. Characterization of these two new desaturases will be of greater value for genetic engineering in industrial production of eicosapentaenoic acid (EPA, C20:5n-3) and docosahexaenoic acid (DHA, C22:6n-3). Due to lack of crystal structure information about n-3 desaturases, for the first time, the view of their predicted structures, binding pockets and substrate tunnels was clearly observed based on molecular docking. This will contribute to strengthening our understanding of the structure-function relationships of n-3 fatty acid desaturases.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors have no conflict of interest.
Figures



Similar articles
-
Use of radiolabeled substrates to determine the desaturase and elongase activities involved in eicosapentaenoic acid and docosahexaenoic acid biosynthesis in the marine microalga Pavlova lutheri.Phytochemistry. 2013 Jun;90:43-9. doi: 10.1016/j.phytochem.2013.02.014. Epub 2013 Mar 22. Phytochemistry. 2013. PMID: 23528573
-
Identification and characterization of new Δ-17 fatty acid desaturases.Appl Microbiol Biotechnol. 2013 Mar;97(5):1973-85. doi: 10.1007/s00253-012-4068-2. Epub 2012 May 27. Appl Microbiol Biotechnol. 2013. PMID: 22639141 Free PMC article.
-
Identification of two novel microalgal enzymes involved in the conversion of the omega3-fatty acid, eicosapentaenoic acid, into docosahexaenoic acid.Biochem J. 2004 Dec 1;384(Pt 2):357-66. doi: 10.1042/BJ20040970. Biochem J. 2004. PMID: 15307817 Free PMC article.
-
The front-end desaturase: structure, function, evolution and biotechnological use.Lipids. 2012 Mar;47(3):227-37. doi: 10.1007/s11745-011-3617-2. Epub 2011 Oct 19. Lipids. 2012. PMID: 22009657 Review.
-
Fatty Acid Desaturases, Polyunsaturated Fatty Acid Regulation, and Biotechnological Advances.Nutrients. 2016 Jan 4;8(1):23. doi: 10.3390/nu8010023. Nutrients. 2016. PMID: 26742061 Free PMC article. Review.
Cited by
-
Molecular mechanism of interaction between fatty acid delta 6 desaturase and acyl-CoA by computational prediction.AMB Express. 2022 Jun 9;12(1):69. doi: 10.1186/s13568-022-01410-0. AMB Express. 2022. PMID: 35680699 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

