The oxazolomycin family: a review of current knowledge
- PMID: 35519217
- PMCID: PMC9057759
- DOI: 10.1039/d0ra08396h
The oxazolomycin family: a review of current knowledge
Abstract
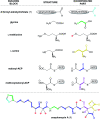
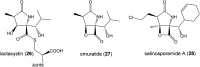
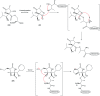
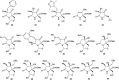
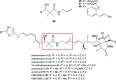
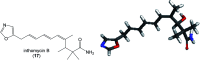
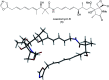
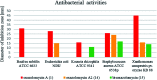
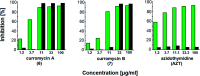
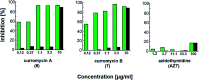
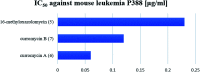
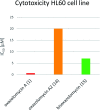
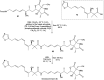
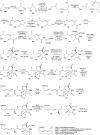
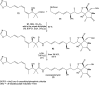
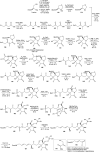
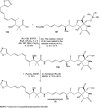
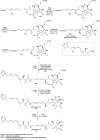
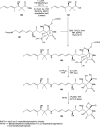
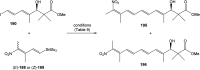
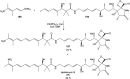
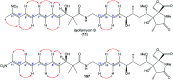
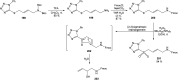
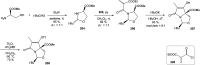
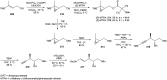
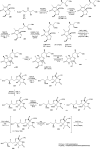
Oxazolomycin A and neooxazolomycin were firstly isolated in 1985 by the group of Uemura et al. from the Streptomyces sp. bacteria. To date, there have been reported 15 different natural compounds commonly classified as part of the oxazolomycin family. All oxazolomycin compounds possess extraordinary structures and they represent a synthetic challenge. Such molecules are additionally known for their wide range of biological activity including antibacterial, antiviral and cytotoxic effects. The present review summarizes the structural elucidation and classification of oxazolomycin compounds, their biosynthesis and biological activity. It is further focused on the total syntheses of oxazolomycins and one formal synthesis reported to date.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures













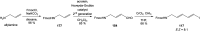
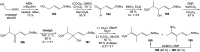
Similar articles
-
Total synthesis of oxazolomycins.Chem Rec. 2014 Aug;14(4):663-77. doi: 10.1002/tcr.201402009. Epub 2014 Jul 28. Chem Rec. 2014. PMID: 25069829 Review.
-
Novel bioactive oxazolomycin isomers produced by Streptomyces albus JA3453.Biosci Biotechnol Biochem. 1998 Mar;62(3):438-42. doi: 10.1271/bbb.62.438. Biosci Biotechnol Biochem. 1998. PMID: 9571773
-
The oxazolomycins: a structurally novel class of bioactive compounds.Curr Drug Discov Technol. 2004 Oct;1(3):181-99. doi: 10.2174/1570163043334974. Curr Drug Discov Technol. 2004. PMID: 16472256
-
Asymmetric Total Synthesis of Oxazolomycins B and C.Chemistry. 2021 Jul 21;27(41):10731-10736. doi: 10.1002/chem.202101341. Epub 2021 Jun 4. Chemistry. 2021. PMID: 33999453
-
Bicyclo [6.3.0] Undecane Sesquiterpenoids: Structures, Biological Activities, and Syntheses.Molecules. 2019 Oct 30;24(21):3912. doi: 10.3390/molecules24213912. Molecules. 2019. PMID: 31671644 Free PMC article. Review.
Cited by
-
Optimized expression of oxazolomycins in engineered Streptomyces longshengensis and their activity evaluation.Microb Cell Fact. 2025 May 20;24(1):114. doi: 10.1186/s12934-025-02726-9. Microb Cell Fact. 2025. PMID: 40394577 Free PMC article.
-
Oxazolomycins produced by Streptomyces glaucus and their cytotoxic activity.RSC Adv. 2021 Oct 28;11(55):35011-35019. doi: 10.1039/d1ra06182h. eCollection 2021 Oct 25. RSC Adv. 2021. PMID: 35494745 Free PMC article.
-
Biosynthesis in Streptomyces sp. NRRL S-1813 and regulation between oxazolomycin A and A2.Braz J Microbiol. 2025 Sep;56(3):1519-1528. doi: 10.1007/s42770-025-01735-5. Epub 2025 Jul 11. Braz J Microbiol. 2025. PMID: 40643887
References
-
- Mori T. Takahashi K. Kashiwabara M. Uemura D. Katayama C. Iwadare S. Shizuri Y. Mitomo R. Nakano F. Matsuzaki A. Tetrahedron Lett. 1985;26:1073–1076. doi: 10.1016/S0040-4039(00)98515-0. - DOI
-
- Takahashi K. Kawabata M. Uemura D. Iwadare S. Mitomo R. Nakano F. Matsuzaki A. Tetrahedron Lett. 1985;26:1077–1078. doi: 10.1016/S0040-4039(00)98516-2. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

