Probing the local conformational flexibility in receptor recognition: mechanistic insight from an atomic-scale investigation
- PMID: 35519308
- PMCID: PMC9064033
- DOI: 10.1039/c9ra01906e
Probing the local conformational flexibility in receptor recognition: mechanistic insight from an atomic-scale investigation
Abstract
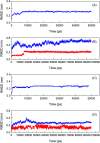
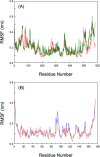
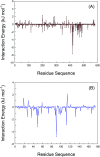
Inherent protein conformational flexibility is important for biomolecular recognition, but this critical property is often neglected in several studies. This event can lead to large deviations in the research results. In the current contribution, we disclose the effects of the local conformational flexibility on receptor recognition by using an atomic-scale computational method. The results indicated that both static and dynamic reaction modes have noticeable differences, and these originated from the structural features of the protein molecules. Dynamic interaction results displayed that the structural stability and conformational flexibility of the proteins had a significant influence on the recognition processes. This point related closely to the characteristics of the flexible loop regions where bixin located within the protein structures. The energy decomposition analyses and circular dichroism results validated the rationality of the recognition studies. More importantly, the conformational and energy changes of some residues around the bixin binding domain were found to be vital to biological reactions. These microscopic findings clarified the nature of the phenomenon that the local conformational flexibility could intervene in receptor recognition. Obviously, this report may provide biophysical evidence for the exploration of the structure-function relationships of the biological receptors in the human body.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Enantioselective recognition of an isomeric ligand by a biomolecule: mechanistic insights into static and dynamic enantiomeric behavior and structural flexibility.Mol Biosyst. 2017 Oct 24;13(11):2226-2234. doi: 10.1039/c7mb00378a. Mol Biosyst. 2017. PMID: 28884187
-
Biological activity of natural flavonoids as impacted by protein flexibility: an example of flavanones.Mol Biosyst. 2015 Apr;11(4):1119-33. doi: 10.1039/c4mb00662c. Mol Biosyst. 2015. PMID: 25673513
-
Estimating the potential toxicity of chiral diclofop-methyl: Mechanistic insight into the enantioselective behavior.Toxicology. 2020 May 30;438:152446. doi: 10.1016/j.tox.2020.152446. Epub 2020 Apr 8. Toxicology. 2020. PMID: 32278049
-
Enzyme dynamics from NMR spectroscopy.Acc Chem Res. 2015 Feb 17;48(2):457-65. doi: 10.1021/ar500340a. Epub 2015 Jan 9. Acc Chem Res. 2015. PMID: 25574774 Free PMC article. Review.
-
Physics of biomolecular recognition and conformational dynamics.Rep Prog Phys. 2021 Dec 8;84(12). doi: 10.1088/1361-6633/ac3800. Rep Prog Phys. 2021. PMID: 34753115 Review.
Cited by
-
Chemoinformatics analysis of Mangifera indica leaves extracted phytochemicals as potential EGFR kinase modulators.Front Chem. 2025 Mar 24;13:1524384. doi: 10.3389/fchem.2025.1524384. eCollection 2025. Front Chem. 2025. PMID: 40196294 Free PMC article.
References
LinkOut - more resources
Full Text Sources

