Detecting early stage structural changes in wild type, pathogenic and non-pathogenic prion variants using Markov state model
- PMID: 35519320
- PMCID: PMC9064127
- DOI: 10.1039/c9ra01507h
Detecting early stage structural changes in wild type, pathogenic and non-pathogenic prion variants using Markov state model
Abstract
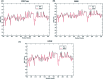
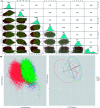
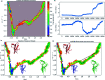
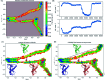
The conversion of prion protein from normal to scrapie followed by the aggregation and deposition of this scrapie form leads to various neurodegenerative diseases. A few studies carried out by researchers suggest that E219K prion mutant (glutamate to lysine mutation at residue position 219) is more stable than wild type protein. However a similar point mutation E200K (glutamate to lysine mutation at residue position 200) is pathogenic. In this study we have carried out detailed atomistic simulation of the wild type, pathogenic mutant E200K and E219K mutant which provides more stability. The aim of the study was to detect the early structural changes present in all the three variants which might be responsible for the stability or for their conversion from PrPC to PrPSc. MSM based analyses have been carried out to find out the differences between WT, E200K and E219K systems. Markov state model (MSM) analysis was able to predict the intermediate states which helped to understand the effect of same mutation at two different locations. The MSM analysis was able to show that the extra stability of E219K mutant may be a result of the increase in number of native contacts, strong salt bridges and less random motions. While pathogenicity of E200K mutant can be attributed to loss of some crucial salt-bridge interactions, increased random motions between helix 2 and helix 3.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
LinkOut - more resources
Full Text Sources

