Exploring the styrene metabolism by aerobic bacterial isolates for the effective management of leachates in an aqueous system
- PMID: 35519756
- PMCID: PMC9055403
- DOI: 10.1039/d0ra03822a
Exploring the styrene metabolism by aerobic bacterial isolates for the effective management of leachates in an aqueous system
Abstract
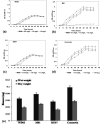
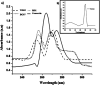
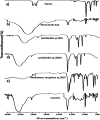
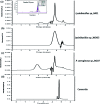
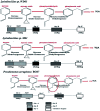
In the present study, the styrene metabolic profile of three aerobic bacterial isolates explored in a batch mode study. The isolates found application in the management of elachates in the waste dump yard. These three bacterial species have different origins and were studied as a single and mixed consortia. The Lysinibacillus strain M01 (from marine sources), Lysinibacillus strain WD03 (from a waste dump yard), and Pseudomonas strain BG07 (from bovine gut) were used in the present study. The styrene concentration was fixed in the range between 0.5 and 1.5 mL L-1. The metabolites obtained upon microbial degradation were assessed using high-performance liquid chromatography (HPLC), UV-visible spectroscopy, and FTIR spectroscopy (Fourier transform infrared spectroscopy). Furthermore, the genes (Sty A, B, C, D, and E) responsible for the degradation of styrene by the three abovementioned isolates were identified using PCR with respective designed primers. Instrumental analyses revealed the presence of phenylacetic acid (PAA) at significant levels in the growth medium after the scheduled experimental period and confirmed the metabolism of styrene by the chosen isolates. Compared to the case of individual cultures, the results of the mixed consortia support the metabolism of styrene at appreciable levels. The present study provides a suitable biological solution for the management of leachates containing styrene and a way to achieve industrially important chemicals (PAA) through a microbially mediated process.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


References
-
- Thaysen C. Stevack K. Ruffolo R. Poirier D. De Frond H. DeVera J. Sheng G. Rochman C. M. Front. Mar. Sci. 2018;5:71. doi: 10.3389/fmars.2018.00071. - DOI
LinkOut - more resources
Full Text Sources

