GhSOC1s Evolve to Respond Differently to the Environmental Cues and Promote Flowering in Partially Independent Ways
- PMID: 35519808
- PMCID: PMC9067242
- DOI: 10.3389/fpls.2022.882946
GhSOC1s Evolve to Respond Differently to the Environmental Cues and Promote Flowering in Partially Independent Ways
Abstract
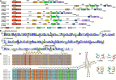
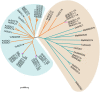
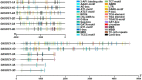
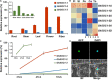
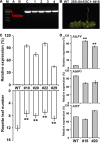
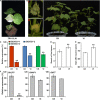
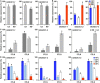
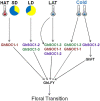
Gossypium hirsutum is most broadly cultivated in the world due to its broader adaptation to the environment and successful breeding of early maturity varieties. However, how cotton responds to environmental cues to adjust flowering time to achieve reproductive success is largely unknown. SOC1 functions as an essential integrator for the endogenous and exogenous signals to maximize reproduction. Thus we identified six SOC1-like genes in Gossypium that clustered into two groups. GhSOC1-1 contained a large intron and clustered with monocot SOC1s, while GhSOC1-2/3 were close to dicot SOC1s. GhSOC1s expression gradually increased during seedling development suggesting their conserved function in promoting flowering, which was supported by the early flowering phenotype of 35S:GhSOC1-1 Arabidopsis lines and the delayed flowering of cotton silencing lines. Furthermore, GhSOC1-1 responded to short-day and high temperature conditions, while GhSOC1-2 responded to long-day conditions. GhSOC1-3 might function to promote flowering in response to low temperature and cold. Taken together, our results demonstrate that GhSOC1s respond differently to light and temperature and act cooperatively to activate GhLFY expression to promote floral transition and enlighten us in cotton adaptation to environment that is helpful in improvement of cotton maturity.
Keywords: Gossypium hirsutum L.; SOC1-like gene; environmental response; evolution; flowering time control.
Copyright © 2022 Ma and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
SEP-like genes of Gossypium hirsutum promote flowering via targeting different loci in a concentration-dependent manner.Front Plant Sci. 2022 Dec 1;13:990221. doi: 10.3389/fpls.2022.990221. eCollection 2022. Front Plant Sci. 2022. PMID: 36531379 Free PMC article.
-
Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L.Plant Cell Rep. 2013 Nov;32(11):1675-86. doi: 10.1007/s00299-013-1479-1. Epub 2013 Jul 27. Plant Cell Rep. 2013. PMID: 23893068
-
Functional characterization of GhSOC1 and GhMADS42 homologs from upland cotton (Gossypium hirsutum L.).Plant Sci. 2016 Jan;242:178-186. doi: 10.1016/j.plantsci.2015.05.001. Epub 2015 May 11. Plant Sci. 2016. PMID: 26566835
-
Genomic organization, differential expression, and functional analysis of the SPL gene family in Gossypium hirsutum.Mol Genet Genomics. 2015 Feb;290(1):115-26. doi: 10.1007/s00438-014-0901-x. Epub 2014 Aug 27. Mol Genet Genomics. 2015. PMID: 25159110
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
Cited by
-
SEP-like genes of Gossypium hirsutum promote flowering via targeting different loci in a concentration-dependent manner.Front Plant Sci. 2022 Dec 1;13:990221. doi: 10.3389/fpls.2022.990221. eCollection 2022. Front Plant Sci. 2022. PMID: 36531379 Free PMC article.
-
GhASHH1.A and GhASHH2.A Improve Tolerance to High and Low Temperatures and Accelerate the Flowering Response to Temperature in Upland Cotton (Gossypium hirsutum).Int J Mol Sci. 2024 Oct 21;25(20):11321. doi: 10.3390/ijms252011321. Int J Mol Sci. 2024. PMID: 39457102 Free PMC article.
-
Comparative Analysis of Floral Transcriptomes in Gossypium hirsutum (Malvaceae).Plants (Basel). 2025 Feb 7;14(4):502. doi: 10.3390/plants14040502. Plants (Basel). 2025. PMID: 40006762 Free PMC article.
-
Genome-wide association study reveals novel SNP loci and candidate genes linked to flowering time in upland cotton.Theor Appl Genet. 2025 Aug 18;138(9):214. doi: 10.1007/s00122-025-05011-w. Theor Appl Genet. 2025. PMID: 40824397
-
Integrated Transcriptomic and Metabolomic Analysis Reveal the Underlying Mechanism of Anthocyanin Biosynthesis in Toona sinensis Leaves.Int J Mol Sci. 2023 Oct 23;24(20):15459. doi: 10.3390/ijms242015459. Int J Mol Sci. 2023. PMID: 37895157 Free PMC article.
References
-
- Amrouk E. M., Mermigkas G., Townsend T. (2021). Recent Trends and Prospects in the World Cotton Market and Policy Developments. Rome: Food and Agriculture Organization of the United Nations, 72.
LinkOut - more resources
Full Text Sources

