A Non-redundant Function of MNS5: A Class I α-1, 2 Mannosidase, in the Regulation of Endoplasmic Reticulum-Associated Degradation of Misfolded Glycoproteins
- PMID: 35519817
- PMCID: PMC9062699
- DOI: 10.3389/fpls.2022.873688
A Non-redundant Function of MNS5: A Class I α-1, 2 Mannosidase, in the Regulation of Endoplasmic Reticulum-Associated Degradation of Misfolded Glycoproteins
Abstract
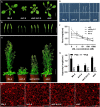
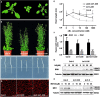
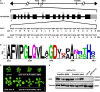
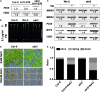
Endoplasmic Reticulum-Associated Degradation (ERAD) is one of the major processes in maintaining protein homeostasis. Class I α-mannosidases MNS4 and MNS5 are involved in the degradation of misfolded variants of the heavily glycosylated proteins, playing an important role for glycan-dependent ERAD in planta. MNS4 and MNS5 reportedly have functional redundancy, meaning that only the loss of both MNS4 and MNS5 shows phenotypes. However, MNS4 is a membrane-associated protein while MNS5 is a soluble protein, and both can localize to the endoplasmic reticulum (ER). Furthermore, MNS4 and MNS5 differentially demannosylate the glycoprotein substrates. Importantly, we found that their gene expression patterns are complemented rather than overlapped. This raises the question of whether they indeed work redundantly, warranting a further investigation. Here, we conducted an exhaustive genetic screen for a suppressor of the bri1-5, a brassinosteroid (BR) receptor mutant with its receptor downregulated by ERAD, and isolated sbi3, a suppressor of bri1-5 mutant named after sbi1 (suppressor of bri1). After genetic mapping together with whole-genome re-sequencing, we identified a point mutation G343E in AT1G27520 (MNS5) in sbi3. Genetic complementation experiments confirmed that sbi3 was a loss-of-function allele of MNS5. In addition, sbi3 suppressed the dwarf phenotype of bri1-235 in the proteasome-independent ERAD pathway and bri1-9 in the proteasome-dependent ERAD pathway. Importantly, sbi3 could only affect BRI1/bri1 with kinase activities such that it restored BR-sensitivities of bri1-5, bri1-9, and bri1-235 but not null bri1. Furthermore, sbi3 was less tolerant to tunicamycin and salt than the wild-type plants. Thus, our study uncovers a non-redundant function of MNS5 in the regulation of ERAD as well as plant growth and ER stress response, highlighting a need of the traditional forward genetic approach to complement the T-DNA or CRISPR-Cas9 systems on gene functional study.
Keywords: BRI1; ERAD; MNS4; MNS5; SBI3.
Copyright © 2022 Sun, Guo, Ali, Zheng, Wei, Zhu, Wang, Li, Li, Zheng, Bai and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Arabidopsis Class I α-Mannosidases MNS4 and MNS5 Are Involved in Endoplasmic Reticulum-Associated Degradation of Misfolded Glycoproteins.Plant Cell. 2014 Apr;26(4):1712-1728. doi: 10.1105/tpc.114.123216. Epub 2014 Apr 15. Plant Cell. 2014. PMID: 24737672 Free PMC article.
-
Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis.Proc Natl Acad Sci U S A. 2011 Jan 11;108(2):870-5. doi: 10.1073/pnas.1013251108. Epub 2010 Dec 27. Proc Natl Acad Sci U S A. 2011. PMID: 21187394 Free PMC article.
-
Unraveling the function of Arabidopsis thaliana OS9 in the endoplasmic reticulum-associated degradation of glycoproteins.Plant Mol Biol. 2012 May;79(1-2):21-33. doi: 10.1007/s11103-012-9891-4. Epub 2012 Feb 11. Plant Mol Biol. 2012. PMID: 22328055 Free PMC article.
-
The Crucial Role of Demannosylating Asparagine-Linked Glycans in ERADicating Misfolded Glycoproteins in the Endoplasmic Reticulum.Front Plant Sci. 2021 Jan 12;11:625033. doi: 10.3389/fpls.2020.625033. eCollection 2020. Front Plant Sci. 2021. PMID: 33510762 Free PMC article. Review.
-
Membrane Protein Quantity Control at the Endoplasmic Reticulum.J Membr Biol. 2017 Aug;250(4):379-392. doi: 10.1007/s00232-016-9931-0. Epub 2016 Oct 14. J Membr Biol. 2017. PMID: 27743014 Free PMC article. Review.
Cited by
-
Protein degrons and degradation: Exploring substrate recognition and pathway selection in plants.Plant Cell. 2024 Sep 3;36(9):3074-3098. doi: 10.1093/plcell/koae141. Plant Cell. 2024. PMID: 38701343 Free PMC article. Review.
-
A Predominant Role of AtEDEM1 in Catalyzing a Rate-Limiting Demannosylation Step of an Arabidopsis Endoplasmic Reticulum-Associated Degradation Process.Front Plant Sci. 2022 Jul 7;13:952246. doi: 10.3389/fpls.2022.952246. eCollection 2022. Front Plant Sci. 2022. PMID: 35874007 Free PMC article.
-
Brassinosteroid Signaling Dynamics: Ubiquitination-Dependent Regulation of Core Signaling Components.Int J Mol Sci. 2025 May 8;26(10):4502. doi: 10.3390/ijms26104502. Int J Mol Sci. 2025. PMID: 40429648 Free PMC article. Review.
-
The Role of SBI2/ALG12/EBS4 in the Regulation of Endoplasmic Reticulum-Associated Degradation (ERAD) Studied by a Null Allele.Int J Mol Sci. 2022 May 22;23(10):5811. doi: 10.3390/ijms23105811. Int J Mol Sci. 2022. PMID: 35628619 Free PMC article.
References
-
- Braun N., Wyrzykowska J., Muller P., David K., Couch D., Perrot-Rechenmann C., et al. (2008). Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20 2746–2762. 10.1105/tpc.108.059048 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

