Combinatorial approach for screening and assessment of multiple therapeutic enzymes from marine isolate Pseudomonas aeruginosa AR01
- PMID: 35519884
- PMCID: PMC9064559
- DOI: 10.1039/c9ra02555c
Combinatorial approach for screening and assessment of multiple therapeutic enzymes from marine isolate Pseudomonas aeruginosa AR01
Abstract
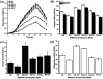
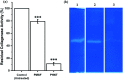
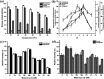
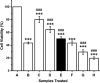
Industrialization and modernization have led to humans being more susceptible to diseases. Therapeutic enzymes from traditional earthbound bacterial origin result have less therapeutic value. Hence, the hunt for a novel source of enzymes is indispensable. Twenty different marine bacterial strains were isolated from mangrove soil around S. P. Pattinum, Tamilnadu, India. From repeated qualitative and quantitative experiments, the study results were that, out of twenty bacterial isolates, only one Gram-negative bacterium was positive for multiple therapeutic enzymes such as asparaginase, glutaminase, uricase and collagenase. Based on its 99% 16S rRNA sequence similarity with Pseudomonas aeruginosa, the isolate was designated as Pseudomonas aeruginosa AR01. Modified minimal medium amended with asparagine results in a simple and cost-effective, one-pot production medium for enhanced production and easy purification of all therapeutic enzymes. The biochemical studies imply that the therapeutic enzymes from P. aeruginosa AR01 may find a significant role in medical applications. The in vitro cytotoxic study reveals that the anticancer enzyme from P. aeruginosa is considerably effective with an IC50 value of 12 μg mL-1 against K-562 cell line. Colony PCR was performed for the detection of specific therapeutic enzyme-coding genes in the genome of P. aeruginosa AR01. PCR results confirm that P. aeruginosa AR01 possesses nucleotide regions for corresponding therapeutic enzymes in its gene cluster. BLASTN and BLASTX analyses of the partial nucleotide sequences of therapeutic enzymes were deposited in GenBank. The results appear so promising that Pseudomonas aeruginosa AR01 may be a potent candidate for medical biotechnology.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Rothia santali sp. nov., endophytic bacteria isolated from sandalwood (Santalum album L.) seedling.Arch Microbiol. 2022 Sep 9;204(10):609. doi: 10.1007/s00203-022-03237-6. Arch Microbiol. 2022. PMID: 36085195
-
Production of a Novel Marine Pseudomonas aeruginosa Recombinant L-Asparaginase: Insight on the Structure and Biochemical Characterization.Mar Biotechnol (NY). 2022 Jun;24(3):599-613. doi: 10.1007/s10126-022-10129-9. Epub 2022 May 4. Mar Biotechnol (NY). 2022. PMID: 35507234
-
Molecular Detection and Phylogenetic Analysis of Pseudomonas aeruginosa Isolated from Some Infected and Healthy Ruminants in Basrah, Iraq.Arch Razi Inst. 2022 Apr 30;77(2):537-544. doi: 10.22092/ARI.2022.357802.2099. eCollection 2022 Apr. Arch Razi Inst. 2022. PMID: 36284961 Free PMC article.
-
Fruit Rot of Tinda Caused by Pseudomonas aeruginosa-A New Report from India.Plant Dis. 2012 Jan;96(1):141. doi: 10.1094/PDIS-05-11-0404. Plant Dis. 2012. PMID: 30731884
-
Quorum quenching activity of Bacillus cereus isolate 30b confers antipathogenic effects in Pseudomonas aeruginosa.Infect Drug Resist. 2019 Jun 7;12:1583-1596. doi: 10.2147/IDR.S182889. eCollection 2019. Infect Drug Resist. 2019. PMID: 31239733 Free PMC article.
Cited by
-
Optimized UV-Spectrophotometric Assay to Screen Bacterial Uricase Activity Using Whole Cell Suspension.Front Microbiol. 2022 Apr 13;13:853735. doi: 10.3389/fmicb.2022.853735. eCollection 2022. Front Microbiol. 2022. PMID: 35495677 Free PMC article.
References
-
- Mane P. Tale V. Int. J. Curr. Microbiol. Appl. Sci. 2015;4(4):17–26.
-
- Gregory A. C. Zayed A. A. Conceicao-Neto N. Temperton B. Bolduc B. Alberti A. et al. . Cell. 2019;117:1–15. - PubMed
LinkOut - more resources
Full Text Sources

