Accelerated trypsin autolysis by affinity polymer templates
- PMID: 35520047
- PMCID: PMC9055874
- DOI: 10.1039/d0ra05827k
Accelerated trypsin autolysis by affinity polymer templates
Abstract
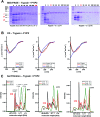
Self-cleavage of proteins is an important natural process that is difficult to control externally. Recently a new mechanism for the accelerated autolysis of trypsin was discovered involving polyanionic template polymers; however it relies on unspecific interactions and is inactive at elevated salt loads. We have now developed affinity copolymers that bind to the surface of proteases by specific recognition of selected amino acid residues. These are highly efficient trypsin inhibitors with low nanomolar IC50 levels and operate at physiological conditions. In this manuscript we show how these affinity copolymers employ the new mechanism of polymer-assisted self-digest (PAS) and act as a template for multiple protease molecules. Their elevated local concentration leads to accelerated autolysis on the accessible surface area and shields complexed areas. The resulting extremely efficient trypsin inhibition was studied by SDS-PAGE, gel filtration, CD, CZE and ESI-MS. We also present a simple theoretical model that simulates most experimental findings and confirms them as a result of multivalency and efficient reversible templating. For the first time, mass spectrometric kinetic analysis of the released peptide fragments gives deeper insight into the underlying mechanism and reveals that polymer-bound trypsin cleaves much more rapidly with low specificity at predominantly uncomplexed surface areas.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
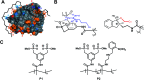


Similar articles
-
Pseudotrypsin: A Little-Known Trypsin Proteoform.Molecules. 2018 Oct 14;23(10):2637. doi: 10.3390/molecules23102637. Molecules. 2018. PMID: 30322187 Free PMC article. Review.
-
Characterization of cold-adapted Atlantic cod (Gadus morhua) trypsin I--kinetic parameters, autolysis and thermal stability.Comp Biochem Physiol B Biochem Mol Biol. 2010 Feb;155(2):186-94. doi: 10.1016/j.cbpb.2009.11.004. Comp Biochem Physiol B Biochem Mol Biol. 2010. PMID: 19913635
-
Recognition of trypsin autolysis products by high-performance liquid chromatography and mass spectrometry.Anal Chem. 1990 Nov 1;62(21):2391-4. doi: 10.1021/ac00220a025. Anal Chem. 1990. PMID: 2291484
-
Evaluation of Pseudotrypsin Cleavage Specificity Towards Proteins by MALDI-TOF Mass Spectrometry.Protein Pept Lett. 2015;22(12):1123-32. doi: 10.2174/0929866522666151008151617. Protein Pept Lett. 2015. PMID: 26446562
-
Getting intimate with trypsin, the leading protease in proteomics.Mass Spectrom Rev. 2013 Nov-Dec;32(6):453-65. doi: 10.1002/mas.21376. Epub 2013 Jun 15. Mass Spectrom Rev. 2013. PMID: 23775586 Review.
References
-
- Hatate H. Toyomizu M. Bull. Jpn. Soc. Sci. Fish. 1985;51:627–633. doi: 10.2331/suisan.51.627. - DOI
-
- Shirdel S. A. Khajeh K. Asghari S. M. Karbalaei-Heidari H. R. Eng. Life Sci. 2014;14:229–234. doi: 10.1002/elsc.201200218. - DOI
-
- Ozturk N. C. Kazan D. Denizci A. A. Erarslan A. Eng. Life Sci. 2012;12:662–671. doi: 10.1002/elsc.201200017. - DOI
LinkOut - more resources
Full Text Sources

