Design and synthesis of novel phenylaminopyrimidines with antiproliferative activity against colorectal cancer
- PMID: 35521305
- PMCID: PMC9066187
- DOI: 10.1039/c9ra03359a
Design and synthesis of novel phenylaminopyrimidines with antiproliferative activity against colorectal cancer
Abstract
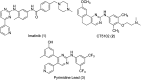
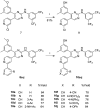
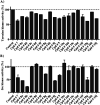
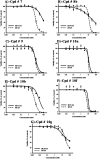
New phenylaminopyrimidine (PAP) derivatives have been designed and synthesised as potential tyrosine kinase inhibitors for the treatment of cancer. The synthesized compounds share a general structure and vary in the substitution pattern at position-2 of the pyridine ring. Several derivatives have demonstrated potent anticancer activities against HCT-116, HT-29 and LS-174T colorectal cancer cells. Furthermore, a number of hits showed good selectivity to Src-kinase. The cytotoxic mechanisms of these compounds were also investigated by studying their effects on cell-cycle distribution. Among all the compounds examined, compound 8b (with a terminal pyridin-3-yl moiety at the pyridine ring) showed the highest inhibitory selectivity towards src-kinase, which was coupled with cell cycle arrest, and apoptotic and autophagic interference, in colorectal cancer cells. This report introduces a novel category of PAP derivatives with promising kinase inhibitory and anticancer effects against colon cancer.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interests.
Figures







Similar articles
-
Structural optimization of 4-(imidazol-5-yl)pyridine derivatives affords broad-spectrum anticancer agents with selective B-RAFV600E/p38α kinase inhibitory activity: Synthesis, in vitro assays and in silico study.Eur J Pharm Sci. 2022 Apr 1;171:106115. doi: 10.1016/j.ejps.2022.106115. Epub 2022 Jan 4. Eur J Pharm Sci. 2022. PMID: 34995782
-
Design, synthesis, and anticancer activity of novel 4-thiazolidinone-phenylaminopyrimidine hybrids.Mol Divers. 2021 May;25(2):1025-1050. doi: 10.1007/s11030-020-10087-1. Epub 2020 Apr 23. Mol Divers. 2021. PMID: 32328961
-
Novel 1,3-diaryl pyrazole derivatives bearing methylsulfonyl moiety: Design, synthesis, molecular docking and dynamics, with dual activities as anti-inflammatory and anticancer agents through selectively targeting COX-2.Bioorg Chem. 2022 Dec;129:106143. doi: 10.1016/j.bioorg.2022.106143. Epub 2022 Sep 11. Bioorg Chem. 2022. PMID: 36191430
-
Synthesis, anticancer activity, toxicity evaluation and molecular docking studies of novel phenylaminopyrimidine-(thio)urea hybrids as potential kinase inhibitors.Comput Biol Chem. 2019 Feb;78:227-241. doi: 10.1016/j.compbiolchem.2018.12.003. Epub 2018 Dec 12. Comput Biol Chem. 2019. PMID: 30579980
-
Progress on the Synthesis of the Aromathecin Family of Compounds: An Overview.Molecules. 2024 May 18;29(10):2380. doi: 10.3390/molecules29102380. Molecules. 2024. PMID: 38792241 Free PMC article. Review.
Cited by
-
2-Chloro-3-cyano-4-nitrobenzyl pyridinium bromide as a potent anti-lung cancer molecule prepared using a single-step solvent-free method.RSC Adv. 2024 Aug 8;14(34):24898-24909. doi: 10.1039/d4ra03538k. eCollection 2024 Aug 5. RSC Adv. 2024. PMID: 39119280 Free PMC article.
-
A Review on Medicinal Approaches of Novel Imatinib Derivatives.Curr Top Med Chem. 2025;25(12):1492-1516. doi: 10.2174/0115680266332163241127114029. Curr Top Med Chem. 2025. PMID: 39779564 Review.
-
Benzochromenopyrimidines: Synthesis, Antiproliferative Activity against Colorectal Cancer and Physicochemical Properties.Molecules. 2022 Nov 15;27(22):7878. doi: 10.3390/molecules27227878. Molecules. 2022. PMID: 36431976 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

