Insights into the biosynthesis pathway of phenolic compounds in microalgae
- PMID: 35521550
- PMCID: PMC9052079
- DOI: 10.1016/j.csbj.2022.04.019
Insights into the biosynthesis pathway of phenolic compounds in microalgae
Abstract
Among the most relevant bioactive molecules family, phenolic compounds (PCs) are well known in higher plants, while their knowledge in microalgae is still scarce. Microalgae represent a novel and promising source of human health benefit compounds to be involved, for instance, in nutraceutical composition. This study aims to investigate the PCs biosynthetic pathway in the microalgal realm, exploring its potential variability over the microalgal biodiversity axis. A multistep in silico analysis was carried out using a selection of core enzymes from the pathway described in land plants. This study explores their presence in ten groups of prokaryotic and eukaryotic microalgae.. Analyses were carried out taking into account a wide selection of algal protein homologs, functional annotation of conserved domains and motifs, and maximum-likelihood tree construction. Results showed that a conserved core of the pathway for PCs biosynthesis is shared horizontally in all microalgae. Conversely, the ability to synthesize some subclasses of phenolics may be restricted to only some microalgal groups (i.e., Chlorophyta) depending on featured enzymes, such as the flavanone naringenin and other related chalcone isomerase dependent compounds.
Keywords: Blue biotechnology; Flavonoids; In silico analysis; Microalgae; Polyphenols.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
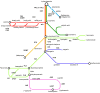
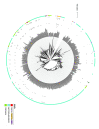
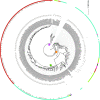
Figures





References
-
- Ancillotti C., Bogani P., Biricolti S., Calistri E., Checchini L., et al. Changes in polyphenol and sugar concentrations in wild type and genetically modified Nicotiana langsdori Weinmann in response to water and heat stress. Plant Physiol Biochem. 2015;97:52–61. - PubMed
-
- Smirnov O.E., Kosyan A.M., Kosyk O.I., Taran N.Y. Response of phenolic metabolism induced by aluminium toxicity in Fagopyrum esculentum moench. plants. Ukr Biochem J. 2015;87:129–135. - PubMed
-
- Handa N., Kohli S.K., Sharma A., Thukral A.K., Bhardwaj R., et al. Selenium modulates dynamics of antioxidative defence expression, photosynthetic attributes and secondary metabolites to mitigate chromium toxicity in Brassica juncea L. plants. Environ Exp Bot. 2019;161:180–192.
-
- Wang J., Yuan B., Huang B. Differential heat-induced changes in phenolic acids associated with genotypic variations in heat tolerance for hard fescue. Crop Sci. 2019;59:667–674.
LinkOut - more resources
Full Text Sources
