Contribution of hydrophobic interactions to protein mechanical stability
- PMID: 35521554
- PMCID: PMC9062142
- DOI: 10.1016/j.csbj.2022.04.025
Contribution of hydrophobic interactions to protein mechanical stability
Abstract
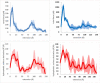
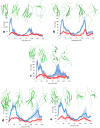
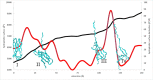
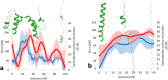
The role of hydrophobic and polar interactions in providing thermodynamic stability to folded proteins has been intensively studied, but the relative contribution of these interactions to the mechanical stability is less explored. We used steered molecular dynamics simulations with constant-velocity pulling to generate force-extension curves of selected protein domains and monitor hydrophobic surface unravelling upon extension. Hydrophobic contribution was found to vary between one fifth and one third of the total force while the rest of the contribution is attributed primarily to hydrogen bonds. Moreover, hydrophobic force peaks were shifted towards larger protein extensions with respect to the force peaks attributed to hydrogen bonds. The higher importance of hydrogen bonds compared to hydrophobic interactions in providing mechanical resistance is in contrast with the relative importance of the hydrophobic interactions in providing thermodynamic stability of proteins. The different contributions of these interactions to the mechanical stability are explained by the steeper free energy dependence of hydrogen bonds compared to hydrophobic interactions on the relative positions of interacting atoms. Comparative analyses for several protein domains revealed that the variation of hydrophobic forces is modest, while the contribution of hydrogen bonds to the force peaks becomes increasingly important for mechanically resistant protein domains.
Keywords: Hydrogen bond; Hydrophobic effect; Protein mechanical stability; Steered molecular dynamics.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Computational and single-molecule force studies of a macro domain protein reveal a key molecular determinant for mechanical stability.Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):1989-94. doi: 10.1073/pnas.0905796107. Epub 2010 Jan 13. Proc Natl Acad Sci U S A. 2010. PMID: 20080695 Free PMC article.
-
Forces stabilizing proteins.FEBS Lett. 2014 Jun 27;588(14):2177-84. doi: 10.1016/j.febslet.2014.05.006. Epub 2014 May 17. FEBS Lett. 2014. PMID: 24846139 Free PMC article. Review.
-
Hydrophobic-hydrophilic forces in protein folding.Biopolymers. 2017 Aug;107(8):10.1002/bip.23020. doi: 10.1002/bip.23020. Biopolymers. 2017. PMID: 28387920 Free PMC article.
-
Molecular dynamics simulations of a beta-hairpin fragment of protein G: balance between side-chain and backbone forces.J Mol Biol. 2000 Mar 3;296(4):1091-104. doi: 10.1006/jmbi.2000.3518. J Mol Biol. 2000. PMID: 10686106
-
Significant role of electrostatic interactions for stabilization of protein assemblies.Adv Biophys. 1997;34:41-54. doi: 10.1016/s0065-227x(97)89630-x. Adv Biophys. 1997. PMID: 9204125 Review.
Cited by
-
A Novel Tri-Hydroxy-Methylated Chalcone Isolated from Chromolaena tacotana with Anti-Cancer Potential Targeting Pro-Survival Proteins.Int J Mol Sci. 2023 Oct 14;24(20):15185. doi: 10.3390/ijms242015185. Int J Mol Sci. 2023. PMID: 37894866 Free PMC article.
-
How strong the interaction really are? Application of nanoITC in the analysis of the interaction between newly synthesized substances with potential anticancer activity and model carrier proteins.Naunyn Schmiedebergs Arch Pharmacol. 2025 Aug;398(8):10111-10124. doi: 10.1007/s00210-025-03884-8. Epub 2025 Feb 13. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 39945815
-
Mining Druggable Sites in Influenza A Hemagglutinin: Binding of the Pinanamine-Based Inhibitor M090.ACS Med Chem Lett. 2024 Nov 28;16(1):126-135. doi: 10.1021/acsmedchemlett.4c00502. eCollection 2025 Jan 9. ACS Med Chem Lett. 2024. PMID: 39811135 Free PMC article.
-
Exploring the antifungal potential of Cannabis sativa-derived stilbenoids and cannabinoids against novel targets through in silico protein interaction profiling.Front Chem. 2025 Jan 6;12:1515424. doi: 10.3389/fchem.2024.1515424. eCollection 2024. Front Chem. 2025. PMID: 39834844 Free PMC article.
-
In Silico Analysis Determining the Binding Interactions of NAD(P)H: Quinone Oxidoreductase 1 and Resveratrol via Docking and Molecular Dynamic Simulations.Eur J Biol. 2023 Dec;82(2):280-288. doi: 10.26650/eurjbiol.2023.1352396. Epub 2023 Nov 23. Eur J Biol. 2023. PMID: 38264080 Free PMC article.
References
LinkOut - more resources
Full Text Sources
