A new artificial intelligence system successfully detects and localises early neoplasia in Barrett's esophagus by using convolutional neural networks
- PMID: 35521666
- PMCID: PMC9278593
- DOI: 10.1002/ueg2.12233
A new artificial intelligence system successfully detects and localises early neoplasia in Barrett's esophagus by using convolutional neural networks
Abstract
Background and aims: Seattle protocol biopsies for Barrett's Esophagus (BE) surveillance are labour intensive with low compliance. Dysplasia detection rates vary, leading to missed lesions. This can potentially be offset with computer aided detection. We have developed convolutional neural networks (CNNs) to identify areas of dysplasia and where to target biopsy.
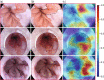
Methods: 119 Videos were collected in high-definition white light and optical chromoendoscopy with i-scan (Pentax Hoya, Japan) imaging in patients with dysplastic and non-dysplastic BE (NDBE). We trained an indirectly supervised CNN to classify images as dysplastic/non-dysplastic using whole video annotations to minimise selection bias and maximise accuracy. The CNN was trained using 148,936 video frames (31 dysplastic patients, 31 NDBE, two normal esophagus), validated on 25,161 images from 11 patient videos and tested on 264 iscan-1 images from 28 dysplastic and 16 NDBE patients which included expert delineations. To localise targeted biopsies/delineations, a second directly supervised CNN was generated based on expert delineations of 94 dysplastic images from 30 patients. This was tested on 86 i-scan one images from 28 dysplastic patients.
Findings: The indirectly supervised CNN achieved a per image sensitivity in the test set of 91%, specificity 79%, area under receiver operator curve of 93% to detect dysplasia. Per-lesion sensitivity was 100%. Mean assessment speed was 48 frames per second (fps). 97% of targeted biopsy predictions matched expert and histological assessment at 56 fps. The artificial intelligence system performed better than six endoscopists.
Interpretation: Our CNNs classify and localise dysplastic Barrett's Esophagus potentially supporting endoscopists during surveillance.
Keywords: AI; Barrett's Esophagus; CAD; CNN; artificial intelligence; computer aided detection; convolutional neural networks; early detection; early neoplasia; neoplasia.
© 2022 The Authors. United European Gastroenterology Journal published by Wiley Periodicals LLC on behalf of United European Gastroenterology.
Conflict of interest statement
MH: Speaker fees (Cook Medical). JGP,DL, DT and PM: Employees at Odin Vision. DS: Share holder Odin vision and Digital Surgery Ltd. LBL: Consultancy and minor share holder Odin Vision. RH: Educational grants to support research infrastructure from Medtronic ltd. Cook endoscopy (fellowship support), Pentax Europe, C2 therapeutics, Beamline diagnostic, Fractyl Ltd.
Figures



Comment in
-
Artificial intelligence in Barrett's oesophagus and the need for shared and combined data.United European Gastroenterol J. 2022 Jul;10(6):525-527. doi: 10.1002/ueg2.12260. Epub 2022 Jun 15. United European Gastroenterol J. 2022. PMID: 35704382 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

