Phenotypic Screening Using High-Content Imaging to Identify Lysosomal pH Modulators in a Neuronal Cell Model
- PMID: 35522480
- PMCID: PMC9121341
- DOI: 10.1021/acschemneuro.1c00804
Phenotypic Screening Using High-Content Imaging to Identify Lysosomal pH Modulators in a Neuronal Cell Model
Abstract
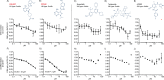
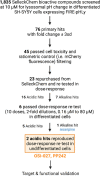
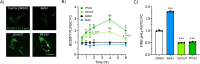
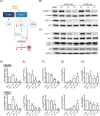
Lysosomes are intracellular organelles responsible for the degradation of diverse macromolecules in a cell. A highly acidic pH is required for the optimal functioning of lysosomal enzymes. Loss of lysosomal intralumenal acidity can disrupt cellular protein homeostasis and is linked to age-related diseases such as neurodegeneration. Using a new robust lysosomal pH biosensor (FIRE-pHLy), we developed a cell-based fluorescence assay for high-throughput screening (HTS) and applied it to differentiated SH-SY5Y neuroblastoma cells. The goal of this study was twofold: (1) to screen for small molecules that acidify lysosomal pH and (2) to identify molecular targets and pathways that regulate lysosomal pH. We conducted a screen of 1835 bioactive compounds with annotated target information to identify lysosomal pH modulators (both acidifiers and alkalinizers). Forty-five compounds passed the initial hit selection criteria, using a combined analysis approach of population-based and object-based data. Twenty-three compounds were retested in dose-response assays and two compounds, OSI-027 and PP242, were identified as top acidifying hits. Overall, data from this phenotypic HTS screen may be used to explore novel regulatory pathways of lysosomal pH regulation. Additionally, OSI-027 and PP242 may serve as useful tool compounds to enable mechanistic studies of autophagy activation and lysosomal acidification as potential therapeutic pathways for neurodegenerative diseases.
Keywords: high-content analysis; lysosomal pH; lysosomes; neurons; pH biosensor; phenotypic screening.
Conflict of interest statement
The authors declare no competing financial interest.
Figures