Machine Learning to Predict Risk of Relapse Using Cytologic Image Markers in Patients With Acute Myeloid Leukemia Posthematopoietic Cell Transplantation
- PMID: 35522898
- PMCID: PMC9126529
- DOI: 10.1200/CCI.21.00156
Machine Learning to Predict Risk of Relapse Using Cytologic Image Markers in Patients With Acute Myeloid Leukemia Posthematopoietic Cell Transplantation
Abstract
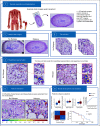
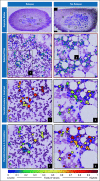
Purpose: Allogenic hematopoietic stem-cell transplant (HCT) is a curative therapy for acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Relapse post-HCT is the most common cause of treatment failure and is associated with a poor prognosis. Pathologist-based visual assessment of aspirate images and the manual myeloblast counting have shown to be predictive of relapse post-HCT. However, this approach is time-intensive and subjective. The premise of this study was to explore whether computer-extracted morphology and texture features from myeloblasts' chromatin patterns could help predict relapse and prognosticate relapse-free survival (RFS) after HCT.
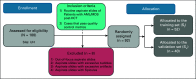
Materials and methods: In this study, Wright-Giemsa-stained post-HCT aspirate images were collected from 92 patients with AML/MDS who were randomly assigned into a training set (St = 52) and a validation set (Sv = 40). First, a deep learning-based model was developed to segment myeloblasts. A total of 214 texture and shape descriptors were then extracted from the segmented myeloblasts on aspirate slide images. A risk score on the basis of texture features of myeloblast chromatin patterns was generated by using the least absolute shrinkage and selection operator with a Cox regression model.
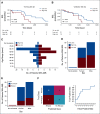
Results: The risk score was associated with RFS in St (hazard ratio = 2.38; 95% CI, 1.4 to 3.95; P = .0008) and Sv (hazard ratio = 1.57; 95% CI, 1.01 to 2.45; P = .044). We also demonstrate that this resulting signature was predictive of AML relapse with an area under the receiver operating characteristic curve of 0.71 within Sv. All the relevant code is available at GitHub.
Conclusion: The texture features extracted from chromatin patterns of myeloblasts can predict post-HCT relapse and prognosticate RFS of patients with AML/MDS.
Conflict of interest statement
Figures
Similar articles
-
Treosulfan, fludarabine, and 2-Gy total body irradiation followed by allogeneic hematopoietic cell transplantation in patients with myelodysplastic syndrome and acute myeloid leukemia.Biol Blood Marrow Transplant. 2014 Apr;20(4):549-55. doi: 10.1016/j.bbmt.2014.01.009. Epub 2014 Jan 16. Biol Blood Marrow Transplant. 2014. PMID: 24440648 Free PMC article. Clinical Trial.
-
Haploidentical Hematopoietic Cell Transplant with Post-Transplant Cyclophosphamide and Peripheral Blood Stem Cell Grafts in Older Adults with Acute Myeloid Leukemia or Myelodysplastic Syndrome.Biol Blood Marrow Transplant. 2017 Oct;23(10):1736-1743. doi: 10.1016/j.bbmt.2017.06.019. Epub 2017 Jul 5. Biol Blood Marrow Transplant. 2017. PMID: 28688919
-
Post-Transplant Cyclophosphamide Combined with Anti-Thymocyte Globulin as Graft-versus-Host Disease Prophylaxis for Allogeneic Hematopoietic Cell Transplantation in High-Risk Acute Myeloid Leukemia and Myelodysplastic Syndrome.Acta Haematol. 2021;144(1):66-73. doi: 10.1159/000507536. Epub 2020 May 19. Acta Haematol. 2021. PMID: 32428903
-
Efficacy of a Second Allogeneic Hematopoietic Cell Transplant in Relapsed Acute Myeloid Leukemia: Results of a Systematic Review and Meta-Analysis.Transplant Cell Ther. 2022 Nov;28(11):767.e1-767.e11. doi: 10.1016/j.jtct.2022.08.008. Epub 2022 Aug 13. Transplant Cell Ther. 2022. PMID: 35970301
-
Generalist in allogeneic hematopoietic stem cell transplantation for MDS or AML: Epigenetic therapy.Front Immunol. 2022 Oct 4;13:1034438. doi: 10.3389/fimmu.2022.1034438. eCollection 2022. Front Immunol. 2022. PMID: 36268012 Free PMC article. Review.
Cited by
-
Survival Prediction of Children Undergoing Hematopoietic Stem Cell Transplantation Using Different Machine Learning Classifiers by Performing Chi-Square Test and Hyperparameter Optimization: A Retrospective Analysis.Comput Math Methods Med. 2022 Sep 25;2022:9391136. doi: 10.1155/2022/9391136. eCollection 2022. Comput Math Methods Med. 2022. PMID: 36199778 Free PMC article.
-
An Automated Pipeline for Differential Cell Counts on Whole-Slide Bone Marrow Aspirate Smears.Mod Pathol. 2023 Feb;36(2):100003. doi: 10.1016/j.modpat.2022.100003. Epub 2023 Jan 9. Mod Pathol. 2023. PMID: 36853796 Free PMC article.
-
Automatic Myeloblast Segmentation in Acute Myeloid Leukemia Images based on Adversarial Feature Learning.Comput Methods Programs Biomed. 2024 Jan;243:107852. doi: 10.1016/j.cmpb.2023.107852. Epub 2023 Oct 18. Comput Methods Programs Biomed. 2024. PMID: 38708372 Free PMC article. No abstract available.
-
Image Analysis in Histopathology and Cytopathology: From Early Days to Current Perspectives.J Imaging. 2024 Oct 14;10(10):252. doi: 10.3390/jimaging10100252. J Imaging. 2024. PMID: 39452415 Free PMC article. Review.
-
The Applications of Machine Learning in the Management of Patients Undergoing Stem Cell Transplantation: Are We Ready?Cancers (Basel). 2025 Jan 25;17(3):395. doi: 10.3390/cancers17030395. Cancers (Basel). 2025. PMID: 39941764 Free PMC article. Review.
References
-
- Swerdlow SH, Campo E, Harris NL, et al. : WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France, IARC Press, 2020
-
- Lin M, Jaitly V, Wang I, et al. : Application of deep learning on predicting prognosis of acute myeloid leukemia with cytogenetics, age , and mutations. arXiv, 1810:1-11, 2018
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

