Quantitative Structure-Activity Relationship (QSAR) Study Predicts Small-Molecule Binding to RNA Structure
- PMID: 35522972
- PMCID: PMC9150105
- DOI: 10.1021/acs.jmedchem.2c00254
Quantitative Structure-Activity Relationship (QSAR) Study Predicts Small-Molecule Binding to RNA Structure
Abstract
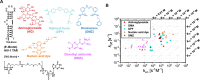
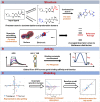
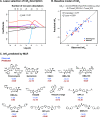
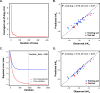
The diversity of RNA structural elements and their documented role in human diseases make RNA an attractive therapeutic target. However, progress in drug discovery and development has been hindered by challenges in the determination of high-resolution RNA structures and a limited understanding of the parameters that drive RNA recognition by small molecules, including a lack of validated quantitative structure-activity relationships (QSARs). Herein, we develop QSAR models that quantitatively predict both thermodynamic- and kinetic-based binding parameters of small molecules and the HIV-1 transactivation response (TAR) RNA model system. Small molecules bearing diverse scaffolds were screened against TAR using surface plasmon resonance. Multiple linear regression (MLR) combined with feature selection afforded robust models that allowed direct interpretation of the properties critical for both binding strength and kinetic rate constants. These models were validated with new molecules, and their accurate performance was confirmed via comparison to ensemble tree methods, supporting the general applicability of this platform.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Ji Q.; Zhang L.; Liu X.; Zhou L.; Wang W.; Han Z.; Sui H.; Tang Y.; Wang Y.; Liu N.; Ren J.; Hou F.; Li Q. Long Non-Coding RNA MALAT1 Promotes Tumour Growth and Metastasis in Colorectal Cancer through Binding to SFPQ and Releasing Oncogene PTBP2 from SFPQ/PTBP2 Complex. Br. J. Cancer 2014, 111, 736–748. 10.1038/bjc.2014.383. - DOI - PMC - PubMed
-
- Gupta R. A.; Shah N.; Wang K. C.; Kim J.; Horlings H. M.; Wong D. J.; Tsai M.-C.; Hung T.; Argani P.; Rinn J. L.; Wang Y.; Brzoska P.; Kong B.; Li R.; West R. B.; van de Vijver M. J.; Sukumar S.; Chang H. Y. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature 2010, 464, 1071–1076. 10.1038/nature08975. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

