Stem-cell-derived trophoblast organoids model human placental development and susceptibility to emerging pathogens
- PMID: 35523141
- PMCID: PMC9136997
- DOI: 10.1016/j.stem.2022.04.004
Stem-cell-derived trophoblast organoids model human placental development and susceptibility to emerging pathogens
Abstract
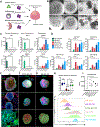
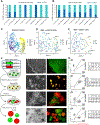
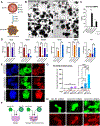
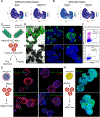
Trophoblast organoids derived from placental villi provide a 3D model system of human placental development, but access to first-trimester tissues is limited. Here, we report that trophoblast stem cells isolated from naive human pluripotent stem cells (hPSCs) can efficiently self-organize into 3D stem-cell-derived trophoblast organoids (SC-TOs) with a villous architecture similar to primary trophoblast organoids. Single-cell transcriptome analysis reveals the presence of distinct cytotrophoblast and syncytiotrophoblast clusters and a small cluster of extravillous trophoblasts, which closely correspond to trophoblast identities in the post-implantation embryo. These organoid cultures display clonal X chromosome inactivation patterns previously described in the human placenta. We further demonstrate that SC-TOs exhibit selective vulnerability to emerging pathogens (SARS-CoV-2 and Zika virus), which correlates with expression levels of their respective entry factors. The generation of trophoblast organoids from naive hPSCs provides an accessible 3D model system of the developing placenta and its susceptibility to emerging pathogens.
Keywords: SARS-CoV-2; X chromosome inactivation; Zika virus; cytotrophoblast; extravillous trophoblast; naive pluripotency; placental development; single-cell transcriptomics; syncytiotrophoblast; trophoblast organoids; trophoblast stem cells.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests I.U.M. serves on the Scientific Advisory Board of Luca Biologics. The Boon laboratory has received unrelated funding support in sponsored research agreements from AI Therapeutics, Green Light Biosciences Inc., and Nano targeting & Therapy BioPharma Inc. The Boon laboratory has received funding support from AbbVie Inc., for the commercial development of SARS-CoV-2 mAb. A.C.M.B. is a recipient of a licensing agreement with AbbVie Inc., for commercial development of SARS-CoV-2 mAb.
Figures

Comment in
-
Taking the guesswork out of journal submission with Community Review.Cell Stem Cell. 2022 May 5;29(5):663-666. doi: 10.1016/j.stem.2022.04.015. Cell Stem Cell. 2022. PMID: 35523136
References
-
- An C, Feng G, Zhang J, Cao S, Wang Y, Wang N, Lu F, Zhou Q, and Wang H (2020). Overcoming Autocrine FGF Signaling-Induced Heterogeneity in Naive Human ESCs Enables Modeling of Random X Chromosome Inactivation. Cell Stem Cell 27, 482–497 e484. - PubMed
-
- Angerer P, Haghverdi L, Buttner M, Theis FJ, Marr C, and Buettner F (2016). destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

