p57Kip2 imposes the reserve stem cell state of gastric chief cells
- PMID: 35523142
- PMCID: PMC9097776
- DOI: 10.1016/j.stem.2022.04.001
p57Kip2 imposes the reserve stem cell state of gastric chief cells
Abstract
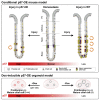
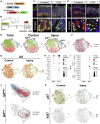
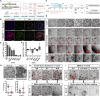
Adult stem cells constantly react to local changes to ensure tissue homeostasis. In the main body of the stomach, chief cells produce digestive enzymes; however, upon injury, they undergo rapid proliferation for prompt tissue regeneration. Here, we identified p57Kip2 (p57) as a molecular switch for the reserve stem cell state of chief cells in mice. During homeostasis, p57 is constantly expressed in chief cells but rapidly diminishes after injury, followed by robust proliferation. Both single-cell RNA sequencing and dox-induced lineage tracing confirmed the sequential loss of p57 and activation of proliferation within the chief cell lineage. In corpus organoids, p57 overexpression induced a long-term reserve stem cell state, accompanied by altered niche requirements and a mature chief cell/secretory phenotype. Following the constitutive expression of p57 in vivo, chief cells showed an impaired injury response. Thus, p57 is a gatekeeper that imposes the reserve stem cell state of chief cells in homeostasis.
Keywords: Gif; Lgr5; Troy; base stem cells; gastric chief cells; p57; reserve stem cells; scRNA-seq; stem cell quiescence; stomach.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Alexa A., Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.36.0. https://bioconductor.org/packages/topGO/
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Ayyaz A., Kumar S., Sangiorgi B., Ghoshal B., Gosio J., Ouladan S., Fink M., Barutcu S., Trcka D., Shen J., et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 2019;569:121–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 212253/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- 203151/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 DK101332/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R37 CA244970/CA/NCI NIH HHS/United States
- I01 BX000930/BX/BLRD VA/United States
- 219478/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- 25636/CRUK_/Cancer Research UK/United Kingdom
- P50 CA236733/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 098357/Z/12/Z /WT_/Wellcome Trust/United Kingdom
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

