Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer
- PMID: 35523176
- PMCID: PMC9197998
- DOI: 10.1016/j.ccell.2022.04.011
Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer
Abstract
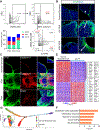
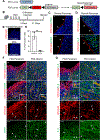
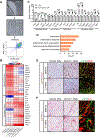
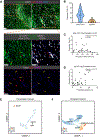
Recent studies have identified a unique cancer-associated fibroblast (CAF) population termed antigen-presenting CAFs (apCAFs), characterized by the expression of major histocompatibility complex class II molecules, suggesting a function in regulating tumor immunity. Here, by integrating multiple single-cell RNA-sequencing studies and performing robust lineage-tracing assays, we find that apCAFs are derived from mesothelial cells. During pancreatic cancer progression, mesothelial cells form apCAFs by downregulating mesothelial features and gaining fibroblastic features, a process induced by interleukin-1 and transforming growth factor β. apCAFs directly ligate and induce naive CD4+ T cells into regulatory T cells (Tregs) in an antigen-specific manner. Moreover, treatment with an antibody targeting the mesothelial cell marker mesothelin can effectively inhibit mesothelial cell to apCAF transition and Treg formation induced by apCAFs. Taken together, our study elucidates how mesothelial cells may contribute to immune evasion in pancreatic cancer and provides insight on strategies to enhance cancer immune therapy.
Keywords: cancer-associated fibroblast; mesothelial cell; mesothelin; pancreatic cancer; regulatory T cell.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
Tracing back to one of the origins of immune evasion in pancreatic cancer.Cancer Cell. 2022 Jun 13;40(6):595-597. doi: 10.1016/j.ccell.2022.05.002. Epub 2022 May 26. Cancer Cell. 2022. PMID: 35623340
References
-
- Barnden MJ, Allison J, Heath WR, and Carbone FR (1998). Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 76, 34–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

