Sars-Cov-2 Spike Protein-Induced Damage of hiPSC-Derived Cardiomyocytes
- PMID: 35523737
- PMCID: PMC9347759
- DOI: 10.1002/adbi.202101327
Sars-Cov-2 Spike Protein-Induced Damage of hiPSC-Derived Cardiomyocytes
Abstract
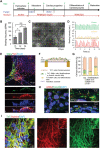
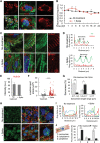
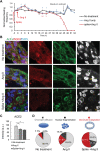
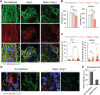
Sars-Cov-2 may trigger molecular and functional alterations of cardiomyocytes (CMs) of the heart due to the presence of receptor angiotensin-converting enzyme 2 (ACE2) of the host cells. While the endocytic itinerary of the virus via cleavage of the spike protein of Sars-Cov-2 is well understood, the role of the remaining part of the spike protein subunit and ACE2 complex is still elusive. Herein, the possible effects of this complex are investigated by using synthetic spike proteins of Sars-Cov-2, human-induced pluripotent stem cells (hiPSC), and a culture device made of an arrayed monolayer of cross-linked nanofibers. hiPSCs are first differentiated into CMs that form cardiac tissue-like constructs with regular beating and expression of both ACE2 and gap junction protein Connexin 43. When incubated with the spike proteins, the hiPSC-CMs undergo a rhythmic fluctuation with overstretched sarcomere structures and dispersed gap junction proteins. When incubated with the spike proteins and supplementary angiotensin II, the damage of the spike protein on hiPSC-CMs is enhanced due to downregulated ACE2, chromatin margination, altered Connexin 43 expression, sarcomere disruption, and beating break. This discovery may imply latent effects of the spike proteins on the heart.
Keywords: ACE2; Ang II; Sars-Cov-2; cardiomyocytes; hiPSC; spike protein.
© 2022 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Wiersinga W. J., Rhodes A., Cheng A. C., Peacock S. J., Prescott H. C., Pathophysiology, T. , JAMA 2020, 324, 782. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

